BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-2327-en.html
2- Department of Microbiology, Karaj Branch, Islamic Azad University, Karaj, Iran
3- Molecular Biology Research Center, Biomedicine Technologies Institute, Baqiyatallah University of Medical Sciences, Tehran, Iran
4- Department of Cell and Molecular Biology, Faculty of Biological Sciences, Kharazmi University, Tehran, Iran
5- Department of Microbiology, Karaj Branch, Islamic Azad University, Karaj, Iran ,
Acinetobacter baumannii (A. baumannii) is a major cause of nosocomial infections. It is often resistant to a wide range of antibiotics, including broad-spectrum antibiotics such as cephalosporins, penicillins, carbapenems, fluoroquinolones, and aminoglycosides. This bacterium can cause pneumonia, bacteremia, meningitis, urinary tract infections, and skin and wound infections through the infected respiratory tract and catheters (1). There are various virulence factors in this bacterium, but one of its most important characteristics is the ability to develop resistance to the various antibiotics (1). This characteristic has increased the prevalence of nosocomial infections caused by this bacterium, especially in intensive care units (ICU), burns, and surgery (2). Mutations, acquisition of new genes, production of drug-destructive enzymes, presence of drug-efflux pumps, and loss of genes encoding purines all play role in the development of broad-spectrum resistance. Due to the rapid acquisition of the resistance genes for different classes of antibiotics, many drugs including penicillins, cephalosporins, tetracyclines, and aminoglycosides have been excluded from the treatment options for the A. baumannii infections (3). Drug efflux systems play a major role in the development of bacterial resistance to different classes of antibiotics. These multicomponent pumps mediate the excretion of toxic compounds, including antibiotics, out of the cell, along with the entry of protons and sodium ions into the cell. Several families of the efflux pumps have been identified in different bacterial species. In A. baumannii, efflux pump-mediated antimicrobial resistance usually is composed of the families; Resistance-Nodulation-cell superfamily Division (RND) and Major Facilitator Superfamily (MFS) (4). AdeIJK, AdeFGH, and AdeABC systems belong to the RND family. The AdeIJK pump can excrete carbapenems, and the AdeFGH system is involved in the development of bacterial resistance to chloramphenicol, clindamycin, and fluoroquinolone (5). The AdeABC pump is the first known system of RND family pumps in A. baumannii, which plays an important excretory system in the development of multidrug-resistant strains (4). The expression of this pump along with the presence of oxacillinases can cause high-level resistance to carbapenems (6).
This system can pump a wide range of antibiotics out of a cell, including aminoglycosides, fluoroquinolones, beta-lactams, tetracycline, erythromycin, chloramphenicol, and trimethoprim (6). The RND family excretory pumps are generally encoded by the chromosomes and catalyze the excretion of each substrate molecule as a proton ion that enters the cell. The RND family is a three-component system consisting of a transporter protein located in the cytoplasmic membrane, a periplasmic protein called Membrane Fusion Protein (MFP), and an Outer Membrane Protein (OMP). Like other RND members, the AdeABC pump consists of three components: AdeA; a membrane fusion protein, AdeB; a 12-fragments cytoplasmic transmembrane, and AdeC; an outer membrane protein. The AdeB transporter protein takes substrates from the phospholipid bilayer of the inner membrane or cytoplasm and then transports them to the extracellular environment via the AdeC membrane protein. The AdeABC pump has been shown to induce antimicrobial resistance in A. baumannii by taking out aminoglycosides, beta-lactams, fluoroquinolones, tetracyclines, macrolides, chloramphenicol, trimethoprim, and ethidium bromide (4). The adeABC operon encodes the AdeABC pump and is tightly controlled by the two-component AdeR-AdeS control, which in turn is encoded by the adeRS operon. The adeRS operon is located upstream of the adeABC genes and is transcribed in the opposite direction. AdeR is a transcriptional regulatory agent and AdeS is a histidine protein kinase sensor located in the cytoplasmic membrane. The AdeS sensor protein monitors environmental conditions and activates or deactivates the response regulator protein that controls pump expression. The phosphorylated response protein is also dephosphorylated by the phosphatase activity of the sensor protein. Histidine kinase enzymes are dual proteins that cause phosphorylation and dephosphorylation of response-regulating factors and regulate their activity (4). Based on several epidemiological reports pointing to the increased rate of nosocomial infections caused by A. baumannii and also the role of antibiotic resistance in increasing the pathogenicity of this bacterium; the possible association between the presence of drug efflux pumps and multidrug resistance were investigated in the clinical isolates of this bacterium.
Collection, Detection, and Identification of Isolates:
This descriptive-analytical study was performed on 120 isolates of A. baumannii isolated from the clinical sources obtained from three hospitals of Milad, Imam Khomeini, and Motahhari in Tehran, Iran for one year. The initial bacterial identification was carried out through culture, various biochemical tests, and molecular methods (1).
Ciprofloxacin Sensitivity Determination by Disk Diffusion:
The susceptibility of A. baumannii isolates to ciprofloxacin (5 μg) paper disks (Mast Co. UK) was determined by Kirby-Bauer disk diffusion method on Müller-Hinton agar medium (Merck) according to the CLSI (2019) guideline. The Escherichia coli ATCC 25922 was used for the quality control of the antibiogram.
Sensitivity Measurement Via MIC Using Micro Broth Dilution Method:
For this purpose, ciprofloxacin dilutions of 0.25-256 μg/mL were prepared in Müller-Hinton broth. Then, from pure culture and 24-hr culture of the bacteria grown in Nutrient agar medium, a microbial suspension with turbidity equivalent to 0.5 McFarland was prepared in a tube containing sterile saline. Then, 297 μL of Müller-Hinton broth, containing different dilutions of the antibiotic, was added into each well. The microbial suspension (3 μL) with turbidity equivalent to 0.5 McFarlane was used as a positive control (culture medium with tested bacteria) and a well containing pure culture medium was designated as a negative control. The microplates were incubated for 18 to 24 hr at 37°C. Finally, the lowest concentration of antibiotic that had no visible turbidity was determined as the MIC.
Effects of CCCP Precipitation on Drug-efflux Pumps in A. baumannii Isolates:
The sensitivity of the isolated bacteria to ciprofloxacin was assessed in the presence of a nonspecific drug pump inhibitor (CCCP) based on the determination of MIC by dilution in Müller-Hinton broth. First, Müller-Hinton broth medium containing 10 µg/mL CCCP was made, and then using this base medium, different dilutions of ciprofloxacin with concentrations of 0.25, 0.5, 1, 2, 4, 8, 16, 32, 64, 128, and 256 µg/mL were prepared. Besides, MHB medium containing CCCP with the mentioned concentration but without ciprofloxacin was prepared to show as a control that this pump inhibitor does not have any antibacterial properties (7). Then, the MIC of ciprofloxacin in the presence of CCCP was calculated using the microdilution broth method. The criterion for the presence of the efflux pump was considered if the MIC of ciprofloxacin in the presence of CCCP in the studied isolates was at least four times lower than the MIC of ciprofloxacin alone (8).
Identification of AdeS, AdeR, and AdeB Genes Using PCR:
Using the PCR method, the presence of adeS, adeR, and adeB genes related to the AdeABC drug efflux pump was investigated in the bacterial isolates. The A. baumannii ATCC 19606 standard strains was used as a positive control. Bacterial genomic DNA was extracted by the boiling method. The primers used to amplify the adeS, adeR, and adeB genes are listed in Table 1. For the samples with negative PCR test, the PCR was repeated a second time to confirm the operation. For each PCR reaction, the following compounds were used. PCR reaction in a final volume of 25 µL that included; 12.5 μL of Color Master Mix (2x) from Sina Gene (1X master mix composition, including Tris-HCl 0.5 M, MgCl2 1.5 mM, dNTPs 0.8 mM and Taq 0.04 Units/μL), 1 μL of each forward and reverse primer, 1 μL target DNA and 9.5 μL of double-distilled water. Each PCR program consisted of 30 replication cycles under the following conditions. Initial denaturation at 94°C for 5 min, 30 thermal cycles including denaturation at 94°C for 60 sec, annealing at 57°C for 60 sec, extension at 72°C for 60 sec, and a final extension at 72°C for 5 min. At the end of the PCR reaction, 6 μl of each amplified product was loaded on 1% agarose gel and then electrophoresed in an electrophoresis tank containing 0.5x TBE buffer for 40 min at V=80 voltage. Finally, the agarose gel was examined for the presence of the desired bands in a UV transilluminator.
Table 1. Primers used to amplify adeB, adeR, adeS genes
| Gene | Primer sequence(5′to3′) | Product size (bp) | Reference |
adeS |
F -TTGGTTAGCCACTGTTATCT R -AGTGGACGTTAGGTCAAGTT |
544 | (9) |
adeR |
F -ACTACGATATTGGCGACATT R -GCGTCAGATTAAGCAAGATT |
447 | (9) |
adeB |
F -TTAACGATAGCGTTGTAACC R -TGAGCAGACAATGGAATAGT |
541 | (9) |
Statistical Analysis:
The SPSS (2019) (SPSS Inc., Chicago, Ill., USA) software and Chi-squared test were used to calculate and analyze the results. Each experiment was repeated three times. The P≤0.05 was considered as an indicator of significant level.
Results of Ciprofloxacin Sensitivity Determination by Disk Diffusion Method:
Determination of ciprofloxacin sensitivity by disk diffusion method on the 120 isolates of A. baumannii showed 108 isolates (90%) resistant, two isolates (1.67%) intermediate, and 10 isolates (8.33%) susceptible (Table 2).
Table 2. Comparison of ciprofloxacin sensitivity with disk diffusion and broth microdilution methods
| MIC Interpretive Criteria (μg/mL) |
Zone Diameter Interpretive Criteria (nearest whole mm) |
Disk Content |
Antimicrobial Agent |
|||||
| Resistant 4 ≤ |
Intermediate 2 |
Susceptible ≤1 |
Resistant ≤15 |
Intermediate 16-20 |
Susceptible 21≤ |
0.5 µg | Ciprofloxacin | |
| 105 | 3 | 12 | 108 | 2 | 10 | Number of isolates (A. baumannii) |
||
Results of MIC for ciprofloxacin:
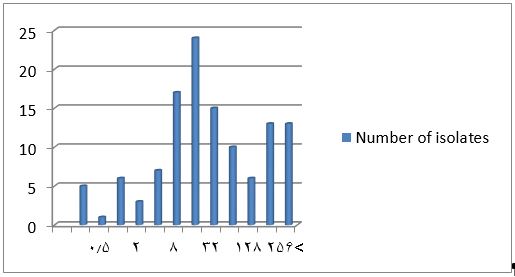
The results showed high resistance levels to ciprofloxacin. For the MIC determination, the susceptibility of the 120 A. baumannii isolates to different concentrations of ciprofloxacin was evaluated by the microdilution method. The results were given in Table 3 and Figure 1. Based on the results, 12 isolates (10%) had MIC of ≤1 μg/mL, which were susceptible to ciprofloxacin, three isolates (2.50%) had a dose of 2 μg/mL, which was intermediate to ciprofloxacin and 105 isolates (87.50 %) had MIC greater than or equal to 4 μg/mL, which were resistant to ciprofloxacin according to the CLSI (2019) definition. This was completely consistent with the results of the disk diffusion method (P≤0.05).
Figure 1. Frequency of different isolates of A. baumannii based on the ciprofloxacin MIC; The horizontal axis shows different concentrations of ciprofloxacin (μg/mL) and the vertical axis shows the number of isolates
Table 3. MIC levels for ciprofloxacin in different isolates of A. baumannii
| Antibiotic | ciprofloxacin (μg/mL) | MIC level of ciprofloxacin (μg/mL) | |||||||||||
| Ciprofloxacin | 0.25-256 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | >256 |
| Number of isolates (A. baumannii) |
5 | 1 | 6 | 3 | 7 | 17 | 24 | 15 | 10 | 6 | 13 | 13 | |
The MIC Levels for Ciprofloxacin in the Presence of 10 μM CCCP:
To determine the role of efflux pump in the resistance to ciprofloxacin, the MIC values in the presence of CCCP, which is a specific inhibitor of the efflux pump, were determined. By definition, if MIC was at least quadrupled in the presence of CCCP, the presence of the drug-efflux pump would be confirmed. The results showed that more than 50% of the total isolates studied in the presence of 10 μM of CCCP reduced the MIC of ciprofloxacin at least twice. However, approximately 47.50% of the isolates that were resistant and semi-sensitive to ciprofloxacin had at least a quadruplicate MIC in the presence of CCCP. Therefore, by definition, at least 47.50% have an active drug-efflux pump (Table 4).
Table 4. The effect of CCCP on ciprofloxacin MIC in A. baumannii isolates
| Number of isolates of A. baumannii | MIC of ciprofloxacin in the absence of CCCP (μg/mL) | MIC of ciprofloxacin in the presence of 10 μM of CCCP (μg/mL) | The amount of reduction MIC of ciprofloxacin in the presence of 10 μM of CCCP (μg/mL) |
| 3 | 2 | 0.5-2 | 0-4 |
| 7 | 4 | 0.5-4 | 0-8 |
| 17 | 8 | 0.25-8 | 0-16 |
| 24 | 16 | 1-16 | 0-16 |
| 15 | 32 | 2-32 | 0-16 |
| 10 | 64 | 8-64 | 0-8 |
| 6 | 128 | 8-128 | 0-16 |
| 13 | 256 | 8-256 | 0-32 |
| 13 | > 256 | 4 - > 256 | 0-64 |
PCR Results of AdeS, AdeR, AdeB Genes:
The presence of genes related to the AdeABC drug-efflux pump in the bacterial isolates was investigated by PCR. For this, the structural gene adeB (transporter gene), and two regulatory genes adeS (sensor kinase gene) and adeR (response regulator gene) were evaluated separately. The results showed that out of the 120 A. baumannii isolates, only three isolates did not have show any band on 1.2% agarose gel for adeB, adeR, and adeS genes. Therefore, based on the results of the PCR test, 98.33% of the isolates had adeB, adeS, and adeR genes or response regulators. The adeB, adeS, and adeR gene amplification responses produced 541, 544, and 447 bp bands, respectively (Figures 2-4).
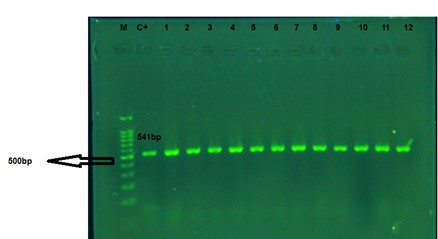
Figure 2. The PCR products of the adeB gene of A. baumannii. M: 100 bp Marker; C+: positive control of A. baumannii ATCC 19606. 1 to 12: the clinical isolates. The amplified fragment size is about 540 bp.
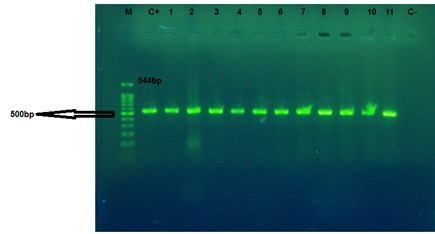
Figure 3. The PCR products of adeS gene of A. baumannii. M: Marker 100bp, C+: positive control of A. baumannii ATCC 19606. 1 to 11: the clinical isolates, and C-: negative control. The amplified fragment size is 544 bp.
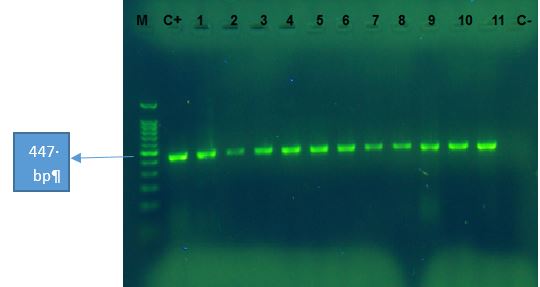
Figure 4. The PCR products of adeR gene for A. baumannii. M: Marker 100bp, C+: positive control of A.baumannii ATCC 19606. 1 to 11: the clinical isolates, and C-: negative control. The amplified fragment size is 447 bp.
This study investigated the presence of AdeABC drug-efflux pumps by a genotypic method as well as the effect of a known chemical efflux pump inhibitor on ciprofloxacin MIC values.
Today, A. baumannii is one of the most common and important nosocomial pathogens isolated from the patients admitted to different wards of hospitals (7). In recent years, this bacterium has become an important cause of death in hospitalized patients, especially in the patients admitted to the ICU wards (8). It seems that the ability of this bacterium to adapt and remain in the hospital environment and its inherent as well as acquired resistance to the most common antibiotics causes their colonization in inpatients and hospital staff and causes nosocomial infections in the hospitalized patients (10, 11). On the other hand, the spread of antibiotic resistance genes and the emergence of multiple antibiotic resistances have made this bacterium a complex problem in developing countries and have caused this bacterium to gain an important place in the clinical microbiology (12). For this reason, this bacterium is known as a very successful pathogen against any treatment and cleansing agents. These unique features have led to the prevention and control of infections caused by this bacterium to become an important health issue in many countries, including Iran (13, 14).
The results of the antibiogram test on the 120 isolates of A. baumannii showed 108 isolates (90%) resistant, two isolates (1.67%) intermediate, and 10 isolates (8.33%) susceptible to ciprofloxacin. The results of MIC by the microdilution method also showed that out of the 120 bacteria studied, 105 isolates (87.50%) were resistant, 3 (2.50%) and 12 (10%) isolates were intermediate and susceptible to ciprofloxacin, respectively. Ciprofloxacin resistance was reported to be 90.9% in the Rahbar et al., study conducted in Tehran from 2005 to 2006 (15). Armin et al., in 2015, found that 94.5% of clinical A. baumannii isolates were resistant to ciprofloxacin (16). In a study conducted by Mirenjad et al., in 2010 in Tehran, antibiotic susceptibility was examined in 50 isolates of A. baumannii and they found 92% of the isolates resistant to ciprofloxacin (17). A study by Aminzadeh et al., during 2006-2009 found that 70% of the isolates were resistant to ciprofloxacin (18). In a study conducted by Peymani et al., in Tabriz, from 2008 to 2009, antibiotic resistance in 110 isolates of A. baumannii was investigated. The antimicrobial resistance to the antibiotic ciprofloxacin was 85% (19). In a study conducted by Basatian-Tashkan et al., in Tehran, 61.6% ciprofloxacin resistance was reported (20). In a study by Mahmoudi et al., the MIC values for ciprofloxacin were determined from 4 to 32 μg/mL and resistance to ciprofloxacin was more than 90% (21). In a study conducted in hospitals of Isfahan city by Ghajavand et al., the resistance to ciprofloxacin in A. baumannii isolates was 100% (22). In a study in Turkey, Baran et al., reported 83.3% resistance to ciprofloxacin in A. baumannii isolates (23). The results of most of these studies were consistent with the results of the current study and showed high resistance to fluoroquinolones.
If the strain isolated from nosocomial infections is sensitive to ciprofloxacin, its clinical application will be better than that of carbapenems (24). On the other hand, many strains of A. baumannii resistant to ciprofloxacin are also resistant to other antibiotics, thus, to treat such infections, other drugs such as carbapenems - sulbactam, colistin, or cyclin should be used. Therefore, looking at resistance levels towards ciprofloxacin will provide sufficient information for the physicians to select appropriate treatment methods against infections caused by this organism (25). Based on the results obtained from the MIC test in the Ardebili study and under reports from other studies, the level of resistance of the most A. baumannii isolates to five different antibiotics; gentamicin, ciprofloxacin, ceftazidime, imipenem, and cefepime were high (24, 26, 27). One of the mechanisms that A. baumannii uses to resist fluoroquinolones, including ciprofloxacin and antibiotics such as tetracycline, is the use of AdeABC transport systems or excretory pumps. Transmission systems are present in all organisms, including eukaryotes, and can introduce or remove various compounds such as physiological substances and antibiotic and non-antibiotic substrates into the cell (4, 6). The bacterial repellent pumps involved in resistance to antimicrobial agents are also important in the bacterial pathogenesis and survival (4). Multidrug resistance due to the overexpression of chromosomal excretion systems in gram-negative bacteria is common and often occurs with the participation of members of the RNA superfamily (28). In A. baumannii, in addition to the acquired resistance indices carried by plasmids, transposons, or islets of resistance, two excretory systems of the RND family; including AdeABC and AdeIJK, play an important role in the development of MDR phenotype. The overexpression of AdeABC by point mutations in its adeR-adeS two-component system or replacement of the ISAbal extension sequence upstream of the adeABC operon coincides with resistance or hypersensitivity aminoglycosides, cefepime, fluoroquinolones, tigecycline, and tetracycline (29). The adeABC operon is inherently present on chromosomes in approximately 80% (from 53% to 97%) of A. baumannii strains (10, 28). This operon is not expressed in A. baumannii isolates and appears to be mainly associated with the clinical isolates. The study of Nemec et al., showed that 82% of A. baumannii isolates (sensitive and antibiotic-resistant) simultaneously have the four genes of adeR, adeS, adeA, and adeB and 35% carry the adeC gene (30, 31). In the study by Huys et al., in Belgium, all 49 strains of multidrug-resistant A. baumannii contained the adeB gene (32). According to the results of the present study, the frequency of the three genes, adeB, adeR, and adeS, was 98.33%, which is consistent with the above studies. These results are almost consistent with the results of the Ardabili et al., study (24). Despite various epidemiological studies on the mechanisms of antibiotic resistance in A. baumannii strains in Iran, the role of drug-repellent pumps in the development of antimicrobial resistance at the phototypic and genotypic levels in this bacterium has not been extensively studied. Therefore, there are no comprehensive statistics on the distribution and frequency of genes associated with excretory pumps, including AdeABC, and how they relate to drug resistance in A. baumannii strains, even at the regional level. Therefore, this issue will be a new perspective for further studies on the prevalence of resistance by drug pumps and methods to prevent and eliminate such mechanisms in strains of A. baumannii and similar bacteria. To evaluate the role of drug excretion systems in bacterial resistance, various compounds called efflux pump inhibitors (EPIs) are widely used in a laboratory. Most repulsion pumps use the Proton Motive Force (PMF) as an energy source to remove substrates from the cell. Compounds such as CCCP and dinitrophenyl, by altering the electrochemical potential of the ultrasonic transmembrane and PMF, can disable bacterial excretory systems. In many studies of CCCP, it has been used to investigate the presence of drug efflux systems in A. baumannii (10). In the Ardebili study, the presence of an active drug pump in 48 isolates of A. baumannii with full and intermediate resistance to ciprofloxacin was evaluated using CCCP with a final concentration of 25 μM/mL. Their results showed that 16 isolates out of a total of 48 isolates (33.3%) have an active ciprofloxacin efflux pump. However, in our study, the use of this concentration and lower concentrations of CCCP caused the death of the studied bacteria. Therefore, a concentration of 10 μM/mL CCCP was used. In a study conducted by Abdi-Ali et al., to evaluate the role of excretory pumps in the drug resistance of A. baumannii isolates, they observed that the MIC level of selected MDR strains decreased by 2 to 4 times in the presence of CCCP (33). In a study conducted by Huang et al., in China, the MIC of 42.8% of carbapenem-resistant A. baumannii was reduced by 4 to 16-fold compared to meropenem in the presence of CCCP (34). In Valentine et al., study, it was shown that the susceptibility of 6 out of 20 isolates of A. baumannii to ciprofloxacin increased 4 to 16 times in the presence of each of the NMP and PAβN inhibitors (9). In our study, it was found that in about 47.50% of the samples, more than 4 times reduction of the ciprofloxacin MIC was detected in the presence of CCCP. In general, according to these findings and the results of the present study, the possibility of the presence of fluoroquinolone excretory systems in A. baumannii isolates and their role in resistance to hypersensitivity to this group of antibiotics is enhanced.
It can be concluded that A. baumannii is present in relatively high frequency in different parts of the hospitals and a large percentage of A. baumannii isolates are resistant to the most common antibiotics and multidrug-resistant strains are constantly increasing. Genes related to the AdeABC drug excretory system are abundant among A. baumannii isolates, which can cause selective antibiotic pressure on microbial strains in hospitals, as well as mutations in excretory systems, causing resistance or hypersensitivity.
The authors would like to express their gratitude to the Dean of the Faculty of Science and the Head of the Microbiology Laboratory of the Islamic Azad University, Karaj Branch.
Ethical Considerations
There is nothing to declare.
Authors’ Contributions
HM and HH designed the topic and wrote the manuscript. HM and HH participated in the initial draft and the revision of the manuscript. HM revised the final version of the manuscript. All authors read and approved the final manuscript.
This study was self-funded.
Conflicts of Interest
The authors declare that they have no competing interests.
Received: 2024/02/14 | Accepted: 2024/05/15 | ePublished: 2024/05/25
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |