BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-2479-en.html
2- Department of Microbiology, Karaj Branch, Islamic Azad University, Karaj, Iran ,
Listeria species are Gram-positive rod-shaped bacteria that are widely distributed in natural habitats. Among different species of this genus, Listeria (L.) monocytogenes is known as an important foodborne pathogen for humans (1–3). The bacterium was first isolated from dead rabbit liver tissue in Sweden and a case of human meningitis in 1918 (4). The unique characteristics of the bacterium including resistance to disinfectants, biofilm formation, tolerance, and proliferation under harsh physicochemical conditions such as low temperature, low pH, and high salt concentration have made it an important pathogen in various food products including unpasteurized dairy products, meat, seafood, and vegetables (2, 5). Food contamination may occur at different stages of the food chain from production and processing to packaging and distribution. Consumption of these products can be dangerous for consumers (6).
Listeria monocytogenes can cause listeriosis, a systemic infection that causes severe complications including sepsis, meningitis, and fetal infection in vulnerable groups including the elderly, pregnant women, infants, and immunocompromised individuals. Although the incidence rate of the disease is low, its mortality rate has been reported to be high (20-30%) (1, 7).
Listeria monocytogenes is able to lyse red blood cells (beta hemolysis) due to the expression of an encoded hemolysin by hly gene called Listeriolysin O. The research shows that the hemolysin expression is necessary for L. monocytogenes virulence. Further investigation of hly gene mutations in the cultured cells showed that pore-forming toxin of listeriolysin O plays a role in the escape of this bacterium from the host cell internal vacuole (4, 8, 9).
Listeria monocytogenes isolates can be grouped into four phylogenetic lineages – lineage I, II, III, and IV. Most isolates of this bacterium associated with human clinical cases belong to the lineages I & II. The strains of lineage II are found in food, while higher incidence rates of human listeriosis are associated with lineage I isolates (10, 11). It seems that lineage II strains are better adapted to the saprophytic and environmental life cycle than lineage I strains (12). Lineage II of L. monocytogenes, due to its lower mutation rate, higher stability in the food processing environment, and its association with raw food materials has a higher probability of survival and persistent contamination in the food chain. These characteristics indicate the importance of precise control of this lineage to prevent food contamination (13).
It was found that the nucleotide sequence differences in several pathogenic genes including Listeriolysin O (hly) gene are associated with phylogenetic groups of L. monocytogenes (14). Since the ability of L. monocytogenes to cause disease may vary based on its phylogenetic groups, their rapid differentiation is very important (13, 14). Phenotypic and DNA-based methods enable differentiation of L. monocytogenes beyond the species and subspecies level (15).
Presently, there are different methods for the diagnosis of L. monocytogenes such as the traditional culture method. This method is a golden standard for the diagnosis, but it is time-consuming due to its need for a long incubation and consequently complex biochemical identification and hemolysis tests (16). Molecular methods have been used to diagnose L. monocytogenes in food and food production environments for many years as an alternative to traditional cultivation methods (17). One of the techniques used is the Mismatch Amplification Mutation Assay-Polymerase Chain Reaction (MAMA-PCR), which is used to classify L. monocytogenes lineage groups based on hly gene (18).
MAMA-PCR is a technique to identify point mutations in the DNA sequences. This method is particularly useful for the rapid screening of mutations in cases where sequencing may be too cumbersome and costly. Therefore, we can use this method to detect point mutations without the need for sequencing. MAMA-PCR is based on principles of PCR in which, amplification of target DNA sequences is mainly performed by oligonucleotide reverse primer with a single mismatch at the 3'-end (19, 20).
This study was designed to identify and classify L. monocytogenes from food samples (including unpasteurized milk, soft cheese, smoked fish, and chicken nuggets) and the environmental samples (water and soil) in Shahriar and Andisheh counties using the MAMA-PCR method. Due to its availability and its high ability to identify point mutations and differentiate different lineages, this method is considered as an effective tool for the epidemiological analysis of this pathogen in the study areas. Shahriar and Andisheh counties were selected for sampling because of their locations, being the suburbs of Tehran, their vicinities to Alborz province, as well as their diverse social context and different ethnicities.
Due to the lack of such studies in the country, the results obtained from the research can fill the knowledge gaps in the field of L. monocytogenes epidemiology on the one hand and improve diagnostic processes, trace the source of contamination, and control disease outbreaks on the other hand. The present research focusing on innovative aspects of fast and precise diagnosis will help to improve the existing knowledge on food and environmental health.
2.1 Selection and Collection of Samples
About 100 environmental and food samples, including 70 food samples (20 raw milk, 20 soft cheese, 10 chive, 10 smoked fish, and 10 chicken nugget) and 30 environmental samples (19 garden soil and 11 well water samples) were randomly collected from different locations in Shahriar and Andisheh counties in 2023 summer and fall.2.2 Enrichment and Cultivation of L. monocytogenes
A Standard strain of L. monocytogenes ATCC 19115 was used as positive control. Two enrichment steps and one cultivation step were performed on a selective solid medium. In the first step of the enrichment, 10 gr of each sample of soft cheese, chive, smoked fish, chicken nuggets, and soil was transferred to 90 ml of Tryptic Soy Broth (TSB) medium. For milk, 10 ml of the sample was added to 90 ml of the TSB medium. Then, the samples were kept in a greenhouse at 37°C for 24 hr. For water samples, 500 ml of each sample was filtered by 0.45 µM Millipore syringe filter and then transferred to TSB medium. In the second step, 0.1 ml of the sample cultured on TSB medium was added to 10 ml of liquid broth culture and kept in the greenhouse at 37°C for 48 hr. Then, the cultivation was performed on PALCAM agar medium. For doing this, a loop was removed from broth culture medium (following 48 hr incubation), cultured on PALCAM agar plate, and put in the greenhouse at 37°C for 48 hr (21, 22).2.3 Identification and Purification of L. monocytogenes from Suspicious Colonies
Suspicious colonies were selected regarding the characteristics of the positive control colony grown on PALCAM agar medium and re-cultured on PALCAM agar medium to separate from other grown colonies (21, 22).2.4. Molecular Characterization by MAMA-PCR
DNA extraction was performed according to the DNA Extraction Kit (Topaz Gene, Tehran, Iran). Using the hly gene sequence in the gene banks and Jinneman and Hill study (14), 6 primers and 5 primer combinations were used (Tables 1 and 2) (a pair of primers for detection of L. monocytogenes, two Forward MAMA primers, and two Reverse MAMA primers to identify lineages).Table 1. Specifications of the selected primers
| Primer | X60035 Location | Primer sequence 5' -> 3' |
| LM4 (for) | 1558-1577 | CAG TTG CAA GCG CTT GGA GT |
| LM5 (rev) | 2003-1984 | CCT CCA GAG TGA TCG ATG TT |
| LMA (for) | 1757-1776 | AAG CCG TAA TTT ACG GTG AC |
| LMB (rev) | 1825-1806 | GTA AGT CTC CGA GGT TGC AA |
| LMC (rev) | 1876-1857 | GAA CTC CTG GTG TTT CTC AA |
| LMD (for) | 1865-1884 | CAC CAG GAG TTC CCA TTG AC |
Table 2. Type of primer combination and specificity to each lineage
| Primer code | Primers | PCR product | Type1 | Type2 | Type3 |
| A | LM4 and LM5 | 446 | + | + | + |
| B | LMA and LM5 | 247 | + | - | - |
| C | LM4 and LMB | 268 | - | - | + |
| D | LM4 and LMC | 319 | - | + | - |
| E | LMD and LM5 | 139 | + | - | - |
The PCR reaction was performed in a final volume of 25 µL, consisting of Master Mix, primers (0.5 µM/each), DNA sample, and double-distilled water (ddH₂O). The amplification process was carried out for 35 cycles using the following thermal cycling conditions: an initial denaturation at 95°C for 10 min, followed by denaturation at 95°C for 30 sec, annealing at 55°C for 1 min, extension at 72°C for 1 min, and a final extension at 72°C for 10 min. PCR products were observed on 1.5% agarose gel.
2.5. Statistical Analysis
3.1. Culture Results and PCR Confirmation for L. monocytogenes Identification
According to the culture results, out of 100 total samples, 36 suspected samples were selected for PCR analysis. Ultimately, the presence of Listeria was confirmed in 22 (22%) samples, including 19 food samples (10 cheese, 6 milk, 1 chive, 1 nugget, and 1 smoked fish) and 3 environmental samples (2 water and 1 soil).
3.2. Identified Lineages of L. monocytogenes
Among 22 L. monocytogenes positive samples, 1 milk sample was classified as Lineage III. Lineage II was identified in 3 milk samples, 8 cheese samples, 1 smoked fish sample, 1 chive sample, 1 nugget sample, and 2 water samples. Additionally, 2 cheese samples, 2 milk samples, and 1 soil sample belonged to no lineage. Figures 1 and 2 represent the results of several examined samples.
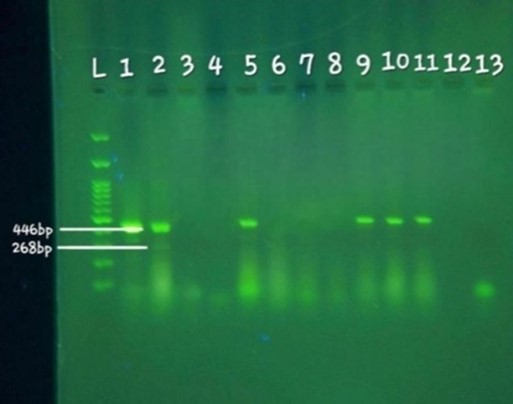
Figure 1. Electrophoresis results of 10 samples examined using MAMA-PCR: L: DNA ladder (100 bp). Well 1: Positive control sample, Listeria monocytogenes reference strain (ATCC 19115). Well 2: Sample related to Lineage III, showing bands at 446 bp (specific to Listeria monocytogenes) and 268 bp (specific to Lineage III). Wells 3-11: Samples examined, showing a band at 319 base pairs (specific to Lineage II) in wells 5, 9, 10, and 11. Well 13: Negative control.
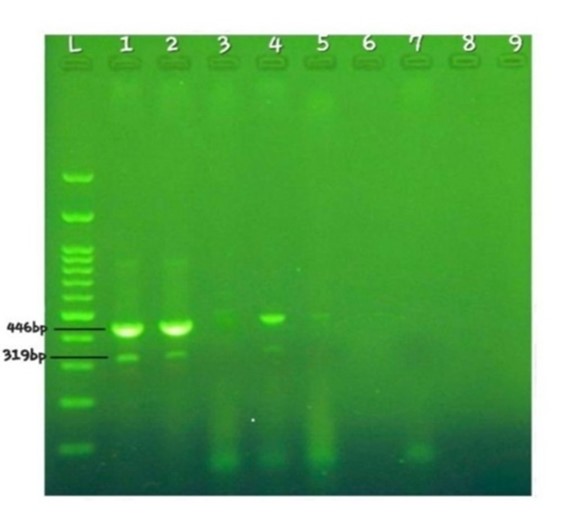
Figure 2. Electrophoresis results of 4 samples examined using MAMA-PCR: L: DNA ladder (100 bp). Well 1: Positive control sample, Listeria monocytogenes reference strain (ATCC 19115). Wells 2-5: Samples examined. Well 7: Negative control. The positive control sample and two other positive samples (wells 2 and 4) exhibit bands at 319 base pairs (specific to lineage II).
3.3 Statistical Analysis of Contamination Frequency
Statistical analyses were conducted using GraphPad Prism software, version 10 (GraphPad Software, USA). Data comparisons were performed using t-test and one-way ANOVA at 95% confidence level with confidence intervals (CI) calculated accordingly. For each statistical test, a hypothesis was considered and P-value less than 0.05 showed the null hypothesis was rejected, indicating significant difference among samples. Asterisks (*,**) represent statistically significant and highly significant differences between groups, respectively.
The results in Table 3 indicated a significant difference in contamination frequency among the samples with L. monocytogenes (P<0.05). Accordingly, cheese and milk samples exhibited the highest levels of contamination, which were significantly different from the other samples. However, no significant difference was observed among chicken nuggets, chives, smoked fish, soil, and water samples. The percentage of contamination in different samples with L. monocytogenes is also shown in Figure 3. The highest contamination was observed in cheese (50%) and milk (30%) samples. This information is visually presented to illustrate the contamination trend in the samples.
Table 3. Investigating the presence of Listeria monocytogenes in the samples and the statistical difference among them.
| Samples | Number | Positive samples (%) | confidence intervals | P-value |
| Cheese | 20 | 10 (45.5) | 0.5 ±0.225 | 0.0156 |
| Milk | 20 | 6 (27) | 0.3 ±0.206 | |
| Nugget | 10 | 1 (4.5) | 0.1 ±0.192 | |
| Chive | 10 | 1 (4.5) | 0.1 ±0.192 | |
| Smoked fish | 10 | 1 (4.5) | 0.1 ±0.192 | |
| Soil | 19 | 1 (4.5) | 0.052 ±0.0989 | |
| Water | 11 | 2 (9.5) | 0.1818 ±0.239 | |
| Total | 100 | 22 (100) |
Significant statistical differences in contamination frequency were determined using Duncan's test.
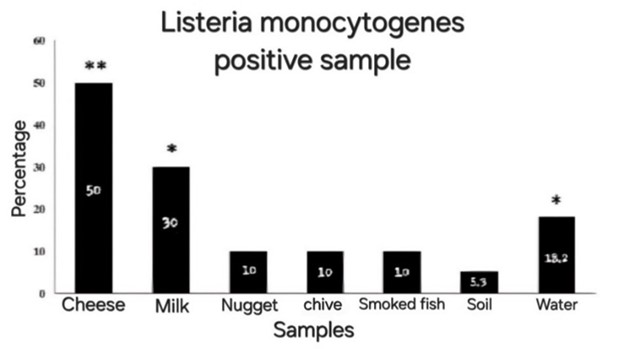
Figure 3. Listeria monocytogenes infection percentage in different samples. The difference marked by * and ** indicates significant and highly significant differences among the samples at the statistical level of 95%.
3.4. Prevalence of L. monocytogenes Lineages
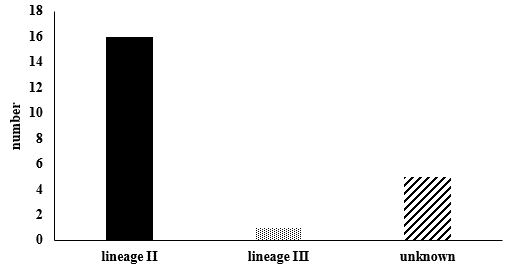
The lineage analysis of the samples using MAMA-PCR revealed that 16 samples (72.8%) belonged to Lineage II, including 8 cheese, 3 milk, 1 chicken nugget, 1 chive, 1 smoked fish, and 2 water samples. In contrast, only 1 milk sample (4.5%) was classified as Lineage III. Additionally, 5 samples (22.7% of the total) could not be assigned to any of the identified lineages (Table 4).
Statistical analysis using the t-test at 95% confidence level demonstrated a significant difference among the lineages of the identified samples (P<0.05) (Figure 4). Moreover, a significant difference was observed between environmental samples (water and soil) and food samples (P<0.05), whereas no significant difference was observed between water and soil samples (P>0.05).
| Samples | Number | Positive samples (%) | Lineage II (%) | Lineage III (%) | Unknown lineage (%) |
| Cheese | 20 | 10 (45.5) | 8 (36.36) | 0 | 2 (9.09) |
| Milk | 20 | 6 (27) | 3 (13.64) | 1 (4.55) | 2 (9.09) |
| Nugget | 10 | 1 (4.5) | 1 (4.55) | 0 | 0 |
| Chive | 10 | 1 (4.5) | 1 (4.55) | 0 | 0 |
| Smoked fish | 10 | 1 (4.5) | 1 (4.55) | 0 | 0 |
| Soil | 19 | 1 (4.5) | 0 | 0 | 1 (4.55) |
| Water | 11 | 2 (9.5) | 2 (9.09) | 0 | 0 |
| Total | 100 | 22 (100) | 16 (72.8) | 1 (4.5) | 5 (22.7) |
| P-value | 0.0246 | ||||
Statistically significant differences in contamination frequency were assessed using Duncan test at 95% confidence level.
Figure 4. Number of specimens found in each lineage. The difference marked by * and ** indicates significant and highly significant differences among the lineages at the statistical level of 95%.
Listeria monocytogenes is a foodborne pathogen that is widely distributed in nature and can enter food processing environments (23). This bacterium is classified into four main phylogenetic lineages. Lineages I and II play a significant role in the occurrence of human diseases, while lineages III and IV are mainly isolated from animal sources and are less present in human and food environments. Lineage II strains are often more stable in food environments due to their high recombination rate and greater ability to adapt to different ecological conditions (24-26).
The dominance of lineage II in food highlights the importance of careful monitoring of this lineage and rapid and sensitive detection of this pathogen for controlling food contamination. The MAMA-PCR method used in this study is an effective tool to classify the L. monocytogenes lineages because of its high accuracy in detecting point mutations and its high speed compared to traditional cultivation and biochemical tests, which often take 24 to 48 hr (13, 16, 18-20).
The results of the study, in which lineage II was known as the dominant lineage in food and environmental samples, are consistent with similar studies conducted in different parts of the world. For example, in the study carried out by Jashari et al., a total of 117 out of 995 tested samples for L. monocytogenes were positive. By examining positive samples in terms of lineage type, lineage II was found in 51.22% of ready-to-eat food isolates, 76.19% of cooked foods, and 72.73% of raw materials (27). Moreover, in the study by Van de Merwe et al., lineage II had the highest ratio of the studied isolates (59.4%), while few genomes belonged to lineages III and IV (13).
This study also investigated the possibility of the presence of L. monocytogenes in several food and environmental samples. Identification of this bacterium in all the samples studied indicates its high importance for the public health. In this study, raw milk and soft cheese had the highest level of contamination among the samples examined. These findings are consistent with the results of Kayode and Okoh in which, L. monocytogenes was found in 42.86% of cheese samples (28). Furthermore, the study carried out by Najafi et al., showed that the prevalence of this lineage in raw milk was 35% (29), which is close to the results of this study.
In the case of smoked fish, the contamination percentage in this study (10%) was higher than in the study by Kester et al., (2.78%) (30). This difference may stem from different sampling methods and various geographical conditions. In the study by Iacumin et al., the percentage of contamination was reported to be 3.4% (31), which is still lower than the current study. The difference may also be due to differences in the sample size and geographic areas under the study.
Regarding vegetables, the study performed by Babazadeh Naseri and Soltan Dallal showed that the contamination rate of the chive with Listeria species was about 25%. But no more details about the L. monocytogenes have been reported (32). In this research, out of 10 chicken nugget samples, only one case was found contaminated. This finding is inconsistent with some previous studies. For example, in 120 chicken-made samples including nuggets studied by Khalafalla et al., no positive case was reported for the presence of L. monocytogenes (33). In contrast, Akbarpour and Bahador, in a study on 40 chicken nuggets, confirmed four L. monocytogenes isolates (34). The differences could be due to the variations in geographical areas, sampling methods, and storage conditions.
The results of the research also showed that the environmental samples (soil and water) can be considered as a source of contamination with this bacterium. Out of 30 environmental samples examined, 2 water and 1 soil samples were contaminated with this bacterium strain. In the study carried out by Linke et al, it was found that 6% of soil samples were contaminated by L. monocytogenes (21). Moreover, the study by Tahir et al., showed the presence of L. monocytogenes in 17.5% of soil samples (35). This rate is higher than that reported in the current study (5.3%) and the reason for this is probably the difference in the sample size and geographic environment under the study.
Raschle et al., identified L. monocytogenes contamination in 13% of 191 water samples, 52% and 48% of the isolates belonged to the lineages I and II, respectively (36). In the current study, both contaminated water samples were classified as lineage II. The difference in sample size and study area may explain this variation.
These findings highlight the need for conducting similar studies in different geographic regions and various food types. Such studies can help assess regional differences in contamination and better inform global food safety practices and risk management strategies.
One of the strengths of this study is diversity of samples and investigation of L. monocytogenes lineages using MAMA-PCR method, which has received little attention in Iran despite the significant role of these lineages in pathogenicity.
However, there are some limitations. For example, confirming the identified lineages using whole genome sequencing could have increased the accuracy of the findings. Moreover, increasing sample size and conducting studies in more diverse geographical areas and sample types could increase the generalizability of the results.
It is recommended that future research investigate the prevalence and distribution of this bacterium over longer observations and wider locations and also pay special attention to identifying the association between L. monocytogenes lineages and clinical cases in vulnerable populations (including elderly and pregnant women). Moreover, evaluating the impact of new molecular identification technologies on improving the diagnosis and control process of the pathogen can play an important role in promoting public health.
This study emphasizes the need for more comprehensive and complementary studies on lineages of L. monocytogenes, especially in different regions and on clinical samples. Furthermore, the exact analysis of different lineages of L. monocytogenes can provide valuable information to identify sources of contamination and prevent disease outbreaks. To prevent food and environmental contamination, it is essential to strengthen the health monitoring systems in preparation and distribution processes, and food supply centers. Moreover, continuous training of personnel on principles of environmental and food hygiene, the development of food safety standards, and the use of new and rapid methods to identify contamination can help reduce the risk of disease outbreaks.
This study is part of a master's thesis in the field of Microbiology, specializing in pathogenic microbes, at the Islamic Azad University, Karaj Branch. Authors would like to express gratitude to the Faculty of Science at the Islamic Azad University, Karaj Branch, for their support and assistance.
Ethical Considerations
This study adhered to the ethical guidelines for the collection and analysis of food and environmental samples. All samples were obtained with proper permissions and in compliance with local regulations. The research prioritized the safety and confidentiality of participants by ensuring that no personal data were collected or disclosed. Potential risks associated with L. monocytogenes were carefully managed, and findings will be communicated transparently to relevant stakeholders, including public health authorities, to promote food safety and public awareness.
Authors’ Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Mahdis mohammadjani and Nasser harzandi. The first draft of the manuscript was written by Azam hadaddi. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
This research received no specific grant from any funding agency in the public, commercial, or not for profit sector.
The authors were not utilized AI Tools.
Received: 2024/12/28 | Accepted: 2025/03/4 | ePublished: 2025/03/30
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |