BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-2340-en.html
2- Department of Microbiology, Faculty of Biological Sciences, Alzahra University, Tehran, Iran ,
3- Department of Bacteriology, Pasteur Institute of Iran, Tehran, Iran
4- Growth and Development Research Center, Tehran University of Medical Sciences, Tehran, Iran & Pediatric Pulmonary Disease and Sleep Medicine Research Center, Pediatric Centre of Excellence, Children's Medical Center, Tehran University of Medical Sciences, Tehran, Iran
Anaerobic bacteria are a group of bacteria that cannot survive in aerobic conditions due to the lack of catalase, superoxide dismutase, and peroxidase enzymes. Fusobacterium, Bacteroides, Porphyromonas, Prevotella, Dialister and Veillonella species are obligate anaerobic bacteria (1-3). Anaerobic bacteria that often enter the body through surgery, wounds, trauma, and food maycause a range of gastrointestinal, blood, skin, and lung infections. Considering the pathogenicity and the role of these bacteria in creating problems related to the human health, identifying this group of bacteria seems essential (2, 4).
Cystic fibrosis (CF) is a lung disease with autosomal recessive inheritance due to the mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Increased secretion of the mucus obstructs the airway and creates an oxygen gradient. The anaerobic bacteria that often enter through the mouth can colonize this mucus. The formation of microbial biofilms in the lungs of these patients protects the lethal effect of oxygen and maintains the survival of bacteria (5). Pseudomonas aeruginosa and Staphylococcus aureus are common pathogens in the CF patients that are isolated through throat swab sampling (6). Both of these bacteria are aerobic and their identification is easily done in clinical laboratories. Generally, throat swabs are not suitable for examining anaerobic bacteria. As the production of sputum in this group of pulmonary patients is done continuously, the excessive volume of sputum helps survival of anaerobic bacteria and make it suitable for the laboratory tests.
The genus Fusobacterium is a group of Gram-negative rod-shaped bacteria that are the normal flora of the mouth, digestive system, and reproductive system of women. Fusobacterium species can cause infection in the adjacent areas. Fusobacterium nucleatum is involved in most oral inflammations such as gingivitis and periodontitis (7). This strain can cause tissue damage by producing butyric acid and increasing the transcription of protease inhibitors. Additionally, some research results have shown that P. aeruginosa can provide anaerobic conditions for the growth of F. nucleatum by consuming dissolved oxygen (8). Dialister genus are anaerobic Gram-negative bacilli from the order of Firmicutes. Some Dialister species are associated with periodontitis, gingivitis, and respiratory tract infections (9). Clinical studies in humans have shown an increase in the abundance of Provetella species in some infections such as periodontitis and bacterial vaginosis (10).
The CF patients are often treated with antibiotics due to the common pathogens of this disease; P. aeruginosa and S. aureus. Long-term use of antibiotics in this group of respiratory patients can lead to the drug resistance. On the other hand, biofilm formation increases the resistance to the antibiotic treatments (11). Some studies have shown that despite the inherent resistance of P. aeruginosa to ampicillin, the inhibition of anaerobic microorganisms by ampicillin can limit the growth of P. aeruginosa (12). Therefore, it seems necessary to determine the type and dose of antibiotic therapy in the treatment of pulmonary pathogens in the CF patients.
The present study investigated the presence or absence and the difference between anaerobic bacteria in the sputum samples of the healthy and CF patients, and also the antibiotic resistance pattern of anaerobic bacteria.
2. 1. Subjects and samples:
The sputum samples were collected from the CF patients (n=50) and the healthy individuals (n=18) between October 2021 and December 2022. Clinically stable people referring to the Children's Medical Center (Tehran, IRan) with CF confirmed by the clinical examinations and the ones who were able to expectorate a sputum sample after obtaining informed consent participated in this study.
Before sputum collection, the mouth and throat were washed with normal saline without any interventions. Immediately after sampling, the samples were quantitatively checked and transferred to the thioglycollate (THIO) broth (HiMedia, M009) culture medium under anaerobic conditions (Gas Pak Type A, Cib Biotech Co). A small volume of sputum sample was separated for the direct microscopic examination with the aim of checking the quality of the sample (13).
2. 2. Isolation and identification by culture-based method:
The sputum samples in THIO broth in an anaerobic jar (90% N2, 5% H2, and 5% CO2) were immediately transferred to the laboratory.. The samples were cultured on one type of non-selective culture medium including Brucella blood (BA) agar (Ibresco, i23026), and three types of selective culture medium including Brain heart infusion (BHI) agar (Ibresco, i23022) with vancomycin, Laked brucella blood agar with kanamycin and vancomycin (LKV) and Brucella blood agar with neomycin and vancomycin (VN) (14). The concentration of vancomycin in BHI agar was 7.5 μg/mL and the concentrations of vancomycin and kanamycin in LKV were 7.5 and 0.1 μg/mL, respectively. The concentrations of neomycin and vancomycin in VN culture medium were 100 and 5 μg/mL, respectively. All agar mediums contain 5% defibrinated sheep blood, 10 μg/mL hemin and 5 μg/mL vitamin K1 (14). Hemin (CAS 16009-13-5), kanamycin (K4000), vancomycin (V2002), and neomycin (N6386) were all purchased from Sigma Chemical Company.
After incubation, if no colony was observed in any of the mentioned cultures, the sputum sample inoculated in THIO was cultured on the same blood agar medium. All inoculated cultures were kept in an anaerobic jar for 7 days at 37ºC until a pure isolate was obtained.
Microscopic and macroscopic investigations including colony morphology, Gram staining, hemolysis, oxygen tolerance, kanamycin (1000 μg, Condalab, 7054), vancomycin (5 μg, Condalab, 7380) and colistin (10 μg, Rosco, 66312) antibiotic disks, 20% bile tolerance, catalase and fluorescence in UV 366 nm were considered for the biochemical identification of the isolates (Table 1) (15, 16).
Table 1. Biochemical properties of isolated anaerobic bacteria (9, 16, 17).
| Gram staining | Vancomycin (5 μg) | Kanamycin (1000 μg) | Colistin (10 μg) | Oxygen tolerance | Bile tolerance | Pigment | |
| Fusobacterium spp. | N | R | S | S | O | V | - |
| Dialister spp. | N | R | S | R | O | - | - |
| Prevotella spp. | N | R | R | V | O | - | +/- |
| Lactobacillus spp. | P | S | V | R | F/A | + | + a / - |
N: Gram-negative bacteria; P: Gram-positive bacteria; R: resistant; S: sensitive; V: variable; O: obligate anaerobic; F: Facultative anaerobic; A: Aerotolerant anaerobe; a: infrequent;
Gram-positive isolates were identified using 16S rRNA sequencing due to the impossibility of identification by biochemical method. DNA extraction from single colonies of Gram-positive isolates was performed using Favorgen (FATGK001) along with lysozyme (Bio Basic, LDB0308) according to the manufacturer's protocol.
Polymerase chain reaction (PCR) using 16S rRNA universal primer (341F: CCTACGGGNGGCWGCAG, and 785R: GACTACHVGGGTATCTAATCC) was considered to identify Gram-positive bacteria from the bacterial colonies. The final volume of 25 µl, including 12.5 µl master mix 2x (Sinaclon, MM2062), 1 µl of each forward and reverse primers, 1 µl DNA, and 9.5 µl distilled water was prepared. The PCR cycling program in thermal cycler (Peqlab-PEQStar) was performed as initial denaturation at 95°C for 5 min, 35 cycles of denaturation at 95°C (40 s), annealing at 55°C (2 min), and elongation at 72°C (1 min), and final extension at 72°C for 7 min (18).
Sequencing of PCR products was done by Pishgam Biotech Company (Tehran, Iran). The obtained sequences were compared with those in the National Center for Biotechnology Information (NCBI) database using the BLAST search tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequences were aligned using MUSCLE. Phylogenetic trees were created with MEGA X software version 10.2.5 using maximum composite likelihood and neighbor-joining methods (19).
The statistical analysis of the isolates identified by both culture-based and molecular-based methods was performed using the Fisher's exact test and GraphPad Prism version 9.3 for Windows (GraphPad Software, Boston, Massachusetts, USA).
2. 4. Antimicrobial susceptibility:
Resistance to ampicillin, clindamycin and ampicillin/sulbactam (Liofilchem, Italy) was determined with E-test method. First, each isolate was cultured in Brucella blood agar containing hemin and vitamin K1 under anaerobic conditions and then a suspension of bacteria with turbidity equal to 1 McFarland was prepared in THIO medium (14). The minimum inhibitory concentration MIC (µg/mL) was recorded based on the breaking point determined by the Clinical and Laboratory Standards Institute (CLSI- M 100, 2020) as resistant, intermediate, and sensitive (20, 21).
2. 5. Ethics statement:
The study was approved by the Research Ethical Committees of Alzahra University (IR.ALZAHRA.REC.1401.006). Informed consent was obtained from all participants and/or their legal guardians.
3. 1. Subjects and samples:
The sputum samples from 68 individuals were analyzed. Of which, 58% were female and 42% were male in the CF group (n=50). In the healthy group (n=18), 16% were female and 83% were male. Out of the 50 CF patients examined, 24 had sputum culture test results using swabs. The culture results of the patients were positive for P. aeruginosa 54.1 % (n=13), S. aureus 37.5 % (n=9), and Klebsiella spp. 8.3 % (n=2). The results of these tests were recorded from the infection detected 8 months before sampling to the time of sampling.
3. 2. Culture-based isolation and identification:
The initial isolation was done without examining the pattern of oxygen tolerance and just by examining the microscopic results. In total, 172 isolates with frequencies of BA=46.51%, BHI=24.41%, LKV=14.53%, and VN= 14.53% were isolated from the CF group and 23 isolates with frequencies of BA=65.21% and BHI=34.78% were isolated from the healthy group. No bacterial isolates were identified in the healthy group using LKV and VN culture media while most bacterial isolates in both groups were isolated using a non-selective BA medium.
Using the Fisher's exact test, no significant relationship was found between Gram-positive (P=0.437) and Gram-negative (P>0.999) isolates separately in the CF and healthy groups. After examining the oxygen tolerance pattern, Gram-negative bacteria were identified by the biochemical analysis and Gram-positive bacteria were identified by the biochemical analysis and 16S rRNA sequencing, the results of which are shown in Table 2.
Table 2. Identification and frequency of anaerobic isolates among healthy and CF patients groups.
| Healthy group (n=18) | CF group (n=50) | |||
| Gram-positive bacteria | Gram-negative bacteria | Gram-positive bacteria | Gram-negative bacteria | |
| Lactobacillus salivarius (2), Lactobacillus rhamnosus (2), |
Dialister spp. (1), Fusobacterium spp. (1), |
Lactobacillus salivarius (1), Lactobacillus rhamnosus (2), Lacticaseibacillus paracasei (1), Lactobacillus spp.(2) |
Dialister spp. (1), Fusobacterium spp. (4), Prevotella spp. (1) |
Bacterial isolates (n) |
| 4 (22/22%) | 2 (11/11%) | 6 (12 %) | 6 (12 %) | Number |
| 6 (33/33%) | 12 (24%) | Overall | ||
3. 3. Molecular identification:
The PCR products from DNA extraction of Gram-positive isolates were sequenced. The phylogeny tree of Gram-positive anaerobic strains isolated using culture and identification by sequencing is shown in Figure 1. All identified Gram-positive isolates belong to the Lactobacillus genus.
3. 4. Antimicrobial susceptibility:
The most Gram-positive and Gram-negative isolates of both healthy and CF groups were sensitive to clindamycin and ampicillin/sulbactam antibiotics. This value was found in Dialister spp. isolates to be intermediate in the healthy (n=1) group and resistant in the CF (n=1) group, respectively. The antibiotic resistance pattern of clinical anaerobic isolates is shown in Table 3.
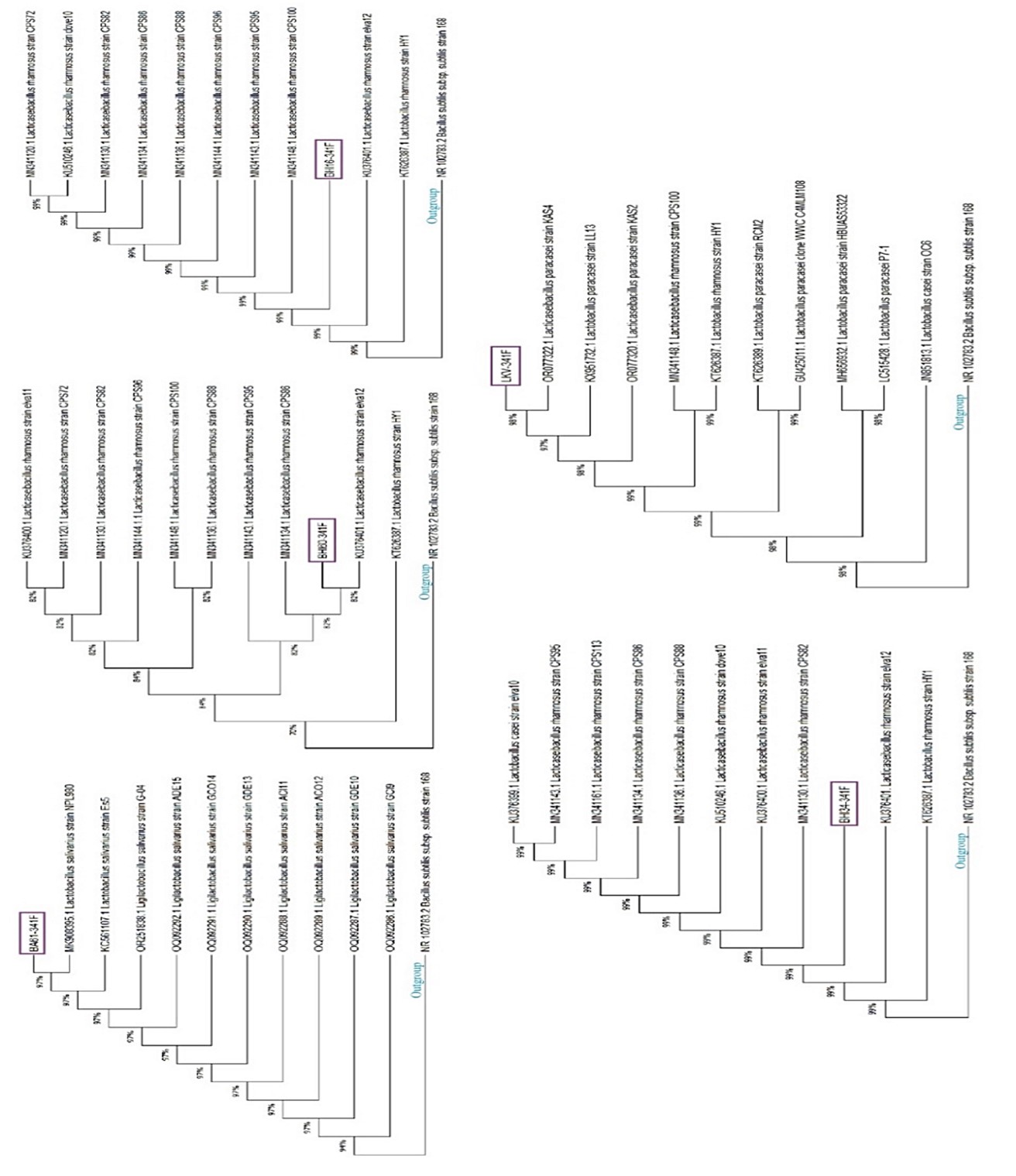
Figure 1. Phylogeny tree of Gram-positive anaerobic strains identified by the sequencing method (16S rRNA). The studied isolates are indicated by purple boxes in each tree. The accession number of each strain in NCBI is mentioned before its name. The trees were rooted by Bacillus subtilis subsp. subtilis strain 168 as the outgroup. The analysis was run in bootstrap 1000.
Table 3. The antibiotic resistance pattern of anaerobic clinical isolates in sputum samples.
| Groups | Clinical isolates | Ampicillin | Ampicillin/sulbactam | Clindamycin |
| Healthy | Fusobacterium spp. | S | S | S |
| Dialister spp. | I | S | S | |
| Lactobacillus rhamnosus | - | - | - | |
| Lactobacillus rhamnosus | I | S | S | |
| Lactobacillus salivarius | S | S | S | |
| Lactobacillus salivarius | S | S | S | |
CF |
Dialister spp. | R | S | S |
| Fusobacterium spp. | I | S | S | |
| Fusobacterium spp. | - | - | - | |
| Fusobacterium spp. | I | S | S | |
| Fusobacterium spp. | R | S | S | |
| Prevotella spp. | - | - | - | |
| Lacticaseibacillus paracasei | I | S | S | |
| Lactobacillus rhamnosus | I | S | S | |
| Lactobacillus rhamnosus | I | S | S | |
| Lactobacillus salivarius | S | S | S | |
| Lactobacillus spp. | S | S | S | |
| Lactobacillus spp. | I | S | S |
S: sensitive, I: intermediate, R: resistant, -: not determined
Sputum production and mucus increase due to mutation in the CFTR gene in the CF patients occurs continuously. The sputum is a rich source of compounds that provide conditions for the growth of microorganisms (22). Common bacterial pathogens in this group of patients are P. aeruginosa and S. aureus (6). For this reason, the experts often pay attention to the results of the throat culture test of these two bacteria when examining the disease process of people.
The results obtained from other research indicate the presence of anaerobic bacteria in the lungs of the CF patients. Fusobacterium, Prevotella, and Veillonella are common anaerobic bacteria reported by other researchers (13).
To date, the beneficial or harmful roles of anaerobic bacteria in the lungs of CF patients is still debated. Different species-dependent effects have been reported in the same bacterial genus (23). Lung microbiome or microbiota changes in different ages of the CF patients indicate microbe-microbe and microbe-host communication (24). According to some reports, some anaerobic bacteria can enhance pathogenicity in the interaction with the primary pathogen without changing the density (25, 26). Therefore, it seems necessary to know the bacterial strains, their pathogenesis, and their effect on the lung function.
The study found that the highest number of anaerobic bacterial isolates were obtained from the BA culture medium without the use of antibiotics that limit the growth of other microorganisms. If the objective is to isolate and identify an unknown microorganism or a collection of bacteria, then the BA culture medium is a suitable option. However, the use of culture media that restricts the growth of other microorganisms using specific antibiotics can speed up the identification of specific and pure isolates. Similar studies recommend the use of a wide and diverse range of specific culture media.
Sheep chocolate agar supplemented with 5%, phenylethyl alcohol agar, McConkey agar, LKV, BHI, anaerobic blood agar, and nutrient agar are several types of these culture media. Although this strategy can isolate more bacterial genera, it does not seem cost-effective. In this research, 3 environments VN, LKV, and BHI were used. No anaerobic bacterial strains were isolated from the VN and LKV culture media in the healthy group. Also, among the three specific culture media considered, the most bacterial strain isolated from the BHI culture medium was in both healthy and CF groups. Nevertheless, it seems that the use of an extended culture approach and types of general and specific culture media are more effective for isolating anaerobic bacterial strains. The presence of anaerobic bacteria in the sputum of CF subjects participating in this study, who often had a stable clinical condition and mild disease, has also been reported by Muhlebach et al., which indicates the relationship between anaerobic bacteria and CF disease conditions (27).
According to the results of the study by Lamoureux et al., the highest percentage of anaerobic bacteria isolated from the CF sputum using the extended culture approach is related to Prevotella and Veillonella bacteria (23). While the largest value for the Gram-negative anaerobic bacteria in the present study was related to Fusobacterium. The difference in the obtained results is probably due to the longer incubation time. Prevotella species require a longer incubation time than Fusobacterium. However, Larsen et al., reported a decrease in Prevotella abundance in the lung microbiota of the patients with asthma and chronic obstructive pulmonary disease (COPD) (10).
Fusobacterium isolates from both healthy and CF groups were sensitive to both clindamycin and ampicillin/sulbactam antibiotics. However, intermediate and resistant pattern to ampicillin was observed in the CF patient group because some patients were under treatment with different antibiotics due to infection with S. aureus and P. aeruginosa. While Fusobacterium isolated from the healthy group was sensitive to ampicillin.
Previously, Moogahi et al. (28), identified anaerobic bacteria and Moazami Goudarzi et al. (29), identified aerobic bacteria in the lungs of CF patients in Iran. In Moogahi et al., research, bronchoalveolar lavage (BAL) and sputum samples were analyzed between September 2014 and March 2022 in SouthWest of Iran, and in Moazami Goudarzi et al., research, pharyngeal swab samples were analyzed in November 2018 and August 2019 in Tehran, Iran. According to these studies, as shown, the sampling time, location, and the method of sample collection in the present study were different from the other two studies. In addition, in these two studies, anaerobic bacteria in CF patients clinical samples were not compared with healthy people. This comparison was assessed in the present study.
Some research results indicate the presence of anaerobic bacteria in the lungs of healthy people (30). The results obtained in this study confirmed the presence of anaerobic bacteria in the sputum samples of healthy people as well. In this study, anaerobic bacterial species were isolated from 33% in the healthy group and 24% in the CF group using culture-based method. This value for the CF group was reported as 69% (30) and 59% (27) in similar studies. Isolation and identification of anaerobic bacterial isolates in the clinical samples is a difficult process, however, obtaining a live and pure bacterial colony allows further investigations in the future. Identification of anaerobic bacteria using molecular methods prepares more quickly, more accurate, and timelyresults (31). However, in molecular identification methods, living and non-living pathogens cannot be distinguished. As we know, some pathogens can cause infection and/or inflammation in a live state, and in some cases, the pathogen does not need to be alive.
The frequency of Gram-positive Lactobacillus bacteria in the healthy group is about two times higher than in the CF group. Therefore, considering the probiotic role of this bacterium, the use of food products containing Lactobacillus may be useful in this group of patients.
Abellan-Schneyder et al., investigated the 16S rRNA primers that are most commonly used in the taxonomic classification of bacteria. Based on the published results, universal primers 341F and 785R were a reasonable choice for the human intestinal samples. Due to the presence of anaerobic bacteria in the intestinal samples, primers 341F and 785R were used in the present study. According to the study of Abellan-Schneyder et al., this pair of primers can identify Lactobacillus, Bifidobacterium, and Atopobium (from the group of anaerobic Gram-positive bacteria) at the genus level (18).
The limitations of this study included continuous application of antibiotics when collecting samples from the patient group and the existence of only one sample collection center. On the other hand, considering the possibility and frequency of microbial biofilm formation in the lungs of CF patients, it should be considered that antibiotic-sensitive planktonic strains can be resistant to the same drug in the biofilm state. This fact is significant in the treatment of the CF patients.
The presence of anaerobic bacteria in the lungs of both healthy people and cystic fibrosis patients has been demonstrated. However, the impact of these bacteria on the severity and progression of the disease as well as their function in the pathogenesis of other microorganisms and the effectiveness of antibiotic treatment remains uncertain.
Fusobacterium isolates were detected more frequently in the CF patients than in the healthy individuals. While the frequency of Lactobacillus in the healthy people was higher than in the CF patients. Drug resistance to ampicillin was observed in Fusobacterium and Dialister isolates of the CF patients. While in the group of healthy people, this pattern was sensitive and intermediate, respectively.
The submitted article is the result of a PhD thesis approved (2021/9/29) at Faculty of Biological Sciences, Alzahra University, Tehran, Iran. We would like to thank the patients and their families, as well as the healthy volunteers who participated in this study.
Ethical Considerations
The study was approved by the Research Ethical Committees of Alzahra University (IR.ALZAHRA.REC.1401.006). Informed consent was obtained from all participants and/or their legal guardians.
Authors’ Contributions
Conceptualization, Hamayeli H, Abdi-Ali A, Attaran B and Modaresi M; funding acquisition, Hamayeli H, Abdi-Ali A and Attaran B; validation, Hamayeli H, Abdi-Ali A, Attaran B and Shafiei M; writing-original draft preparation, Hamayeli H; writing—review and editing, Hamayeli H, Abdi-Ali A, Attaran B, Shafiei M and Modaresi M; project administration, Hamayeli H, Abdi-Ali A and Attaran B. All authors have read and agreed to the published version of the manuscript.
This study was supported by grants from the Department of Microbiology, Faculty of Biological Sciences, Alzahra University, Tehran, Iran.
Conflicts of Interest
The authors declare that they have no competing interests.
Received: 2024/02/9 | Accepted: 2024/05/8 | ePublished: 2024/05/25
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |








