BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1763-en.html


 , Davoud Esmaeili2
, Davoud Esmaeili2 

 , Mohammad Mehdi Moghani Bashi1
, Mohammad Mehdi Moghani Bashi1 

 , Mohamad Reiszadeh3
, Mohamad Reiszadeh3 

 , Sirous Naeimi1
, Sirous Naeimi1 


2- Department of Microbiology and Applied Virology Research Center, Baqiyatallah University of Medical Sciences, Tehran, Iran ,
3- Associate Professor of Surgery, School of Medicine, Trauma Research Center, Bqiyatallah al-Azam Hospital, Baqiyatallah University of Medical sciences, Tehran, Iran
Gastric cancer is the 5th most common cancer worldwide. Recent PCR and metagenomic findings indicate that the stomach is inhabited by a powerful microbiota. Radiotherapy, chemotherapy, and surgery have limited effectiveness in treating Gastric cancer. So, it is essential to develop new therapeutic strategies. Cancer prevention, oncogenesis, and chemotherapy effectiveness are influenced by the intestinal microbiota. There's some evidence that certain bacteria can boost the effectiveness of some traditional immunotherapies and antineoplastics (1, 2).
Surgery, chemotherapy, and radiotherapy are currently the most commonly used treatments for cancer. Cancer treatment requires a breakthrough approach. Several anticancer properties have been demonstrated in vitro for bacteriocin, an antimicrobial peptide. As a result, the patient is now looking for alternatives to conventional treatments, which do not exhibit any additional side effects (3).
Bacteriocin is most well-known for its anti-cancer and therapeutic properties (4). The anti-cancer effects of bacteriocins against cancer cells have been mentioned several times (5). Bacteriocins have the potential to inhibit DNA synthesis and membrane proteins that induce apoptosis and cytotoxicity in tumor cells as a cancer treatment (6).
Bacteriocins (cationic) have been shown to increase the cytotoxicity of cancer cells (anionic) when they bind selectively to cancer cells (cationic). By communicating with the cell surface, bacteriocin peptides cause necrosis, killing the tumor-forming cells without harming healthy cells (7). They disrupt membrane integrity and cause cancer cell apoptosis. Furthermore, angiogenesis is inhibited, and cancer is prevented from progressing as a result (8). Apoptosis is evolutionarily conserved and removes damaged and dysfunctional cells. Apoptotic cells are distinguished from healthy cells by markers on their surfaces. One of the lipids that make up biological membranes is phosphatidylcholine (PC), followed by phosphatidylserine (PS) (9).
Various cell lines were examined independently for the effects of recombinant fusion genes containing peptide sequences with anticancer activity and their specific ligands, in addition to R-type Pyocin, Lactocin, and enterocin bacteriocins. Results indicate that treating gastric cancer cells using recombinant protein causes apoptosis at an 80µg/ml concentration (10). In order to evaluate the combined efficacy of bacteriocins on gastric cancer cells, an anticancer protein combination containing protein fusion, Pyocin type R, Enterocin A, and Lactocin containing a specific ligand (AGS) was investigated.
In the initial stage, we designed a recombinant gene cassette that contained Enterocin A, Lactocin, R-type Pyocin, anti-cancer, and the specific ligand gene sequences. Then, sequences of the EntA (AN: CP012522), PynR (AN: AP014651), and Lac genes (AN: X79889) were obtained from the NCBI website. These sequences were connected to each other by linkers. Based on AGS cell line-specific ligand, BDB software was designed. Also, an anti-cancer sequence was added. IACP web-based software was used to test the anti-cancer properties of the recombinant construct sequence.
Using the online gene script software, we analyzed wild-type (WT) genes from Pseudomonas aeruginosa, Enterococcus faecium, and Lactobacillus sakei. Jcat software was then used to replace the main sequence codons of these genes with those that were preferable for expression in the E. coli host due to the fact that their codons were unsuitable for expression in the host. As part of this process, we reexamined the optimized gene using the GeneScript online program. Bioinformatics tools, such as mfold, can be used to analyze the secondary structure of mRNA.
Using ProtParam ExPASy, the physicochemical properties of fusion peptides, such as protein stability indexes, protein half-life, molecular weight, positive amino acids, mean hydrophobicity (GRAVY), and isoelectric point (PI), were determined. For evaluating and analyzing the secondary structural features of the recombinant protein, GOR4 (Garnier–Osguthorpe–Robson) was used. Using the I-TASSER (Iterative Threading ASSEmbly Refinement), the three-dimensional structure of the recombinant protein was also determined.
Lastly, in the gene cassette design process, the ECORΙ enzyme site was considered in section 5ʹ and the XhoΙ enzyme site was considered in section 3ʹ, respectively. There has been an order placed at Biomatik Corp for a specific sequence of EntA-PynR-Lac-ligand fusion gene for the purpose of synthesizing and cloning into the pET22b expression vector in order to fight cancer.
In terms of cloning and expression of recombinant proteins in E. coli, the pET system is the most powerful available. The IPTG-induced pET systems produced T7 RNA polymerase, and this vector's target gene was then transcribed with this RNA polymerase. Then the transcribed RNA gets translated into protein. E. coli BL-21 (DE3) competence cells were prepared. We used heat shock to transfer the recombinant vector into susceptible cells.
It was found that the following procedures were used in order to induce recombinant plasmid synthesis and produce a recombinant fusion protein in E. coli BL21:
One hour after IPTG was added to the culture medium (1-5 hours), we collected 1 ml of samples from the culture medium and placed them in clean microtubes. Around-the-clock induced samples were also prepared. As soon as the precipitates were centrifuged, they were kept at -20°C till the SDS-PAGE process was initiated, which would last for two days. SDS-PAGE was employed to check the expression of the recombinant protein. In order to achieve protein separation, virtually all methods use polyacrylamide. Separation gel solution with a concentration of 12% and a volume of 20 ml, as well as stacking gel solution with a concentration of 4% and a volume of 10 mL, were prepared using this method (see Table 1). After the polymerization of the gels, the samples collected from the induction stage were removed from the freezer, and after melting, 100 mL of 1x sample buffer was added to them. The microtubes were placed in a container containing boiling water for 5 minutes. By programming the power supply, the electrophoresis started with the samples loaded. Run conditions and times were approximate (voltage: 200 V; expected gel current: initially 35-50 mA and finally 20-31 mA; running time: 30-40 minutes). The run was stopped once bromophenol blue had reached the bottom of the gel. Finally, the separating gel was stained with Coomassie-R250 dye.
Table1. Recipes for stacking and separation gels
| Stacking Gel (4%) | Separation Gel (12%) | |
| 30% acrylamide/bis | 1320 µL | 8000 µL |
| 0.5M Tris-Hcl,PH6.8 | 2520 µL | - |
| 0.5M Tris-Hcl,PH8.8 | - | 5000 µL |
| 10% SDS | 100 µL | 200 µL |
| diH2O | 6000 µL | 6700 µL |
| 10% APS | 50 µL | 100 µL |
| TEMED | 20 µL | 20 µL |
| Total volume | 10 ml | 20 ml |
A nickel column chromatography method with histidine tags was used to purify the recombinant proteins (11). There are a lot of physical ways to lyse cells, including ultrasound. This method uses ultrasonic waves to destroy the cells by destroying them with the help of the waves. In the suspension, the cells are lysed. After preparation of the precipitate, it was sonicated in bulk cytolysis buffer (0.5 M NaCl, 20 mM Tris-HCl, 0.005% PMSF, 0.3% TritonX100, 10 mM Imidazole, pH 8.0). Then, ten 40-second cycles at 4°C were used to break the material using a sonicator. Afterward, the sonicated sample was centrifuged for 20 minutes at 12,000 rpm and purified on a column with the supernatant.
Supernatant injection, washing, and isolation of the desired protein will follow. By using a wash buffer (0.5 M NaCl, 20 mM Tris, 20 mM Imidazole, pH 8.0), the column was cleaned. Through the separation buffer (0.5 M NaCl, 20 mM Tris, 0.6 M Imidazole, pH 8.0), the histidine sequence recombinant proteins were removed from the resin. Recombinant proteins contain histidine sequences, which allow them to bind to metal ions on resin, and the imidazole gradient separated the recombinant protein bound to nickel. In the end, samples collected from the column at different stages of purification were assayed by SDS-PAGE.
As well as Western blots (immunoblots), the antigenicity of the fusion protein was assessed by immunoblot. An immunoblotting method with a goat anti-mouse peroxidase conjugate and anti-his tag monoclonal antibody was used (12). Several blocking substances are used to block the PVDF paper background in order to begin immunoblotting. For this research, Tween 20 was used at a concentration of 0.05-0.1%. T-TBS buffer was then used to wash the membrane. As a next step, the membrane was placed in a buffer containing an anti-his tag monoclonal antibody diluted by 0.5 µg/ml and then incubated for 1-2 hours with a labeled secondary antibody (Goat anti-mouse - Peroxidase conjugate) diluted by T-TBS. TBS buffer was then used to wash the membrane. It was then necessary to expose the membrane to a sufficient amount of solution containing the peroxidase substrate after this step. In order to do this properly, it should be done in the dark. The bands usually appear 5 to 15 minutes after the announcement.
2-1- Cell treatment and study of apoptosis
An incubator containing CO2 was used to incubate the 96-well plate with 100,000 AGS cells from the Genetic and Biological Resource Center of Iran (IBRC C10071) for 24 hours at 37°C. The proliferation of cells and adhesion to the cavity bottom were observed using a microscope. It was possible to prepare concentrations of recombinant protein between 10 and 100 µg/ml at intervals of 10. It took 24 hours for the test to incubate, and then an ELISA reader measured Optical Density (OD). According to the control OD (C), the OD of each concentration (T) was determined. A formula for the percentage of lethality was used to calculate the OD of each concentration (T) and control (C) to calculate the amount of lethality (percentage) in each concentration: %Cytotoxicity=1-T/C *100
In order to determine the extent of apoptosis, cells adhered to the bottom of a 24-well cell culture plate after 24 hours. Several control wells were injected with freshly prepared recombinant protein 80 µg/ml in three wells of the plates (in triplicate). In the end, 1 cc was added by the culture medium. As a result, a CO2 incubator was used to incubate the cell plate for 24 hours at 37°C.
The bottom of each plate was treated with trypsin after the supernatant fluid was removed and washed with PBS. Using an inverted microscope, the separation of the cells was examined. We then placed the treated and untreated cells into a centrifuge tube, applied 30 microliters of recombinant protein at a concentration of 80 microliters per ml, and then centrifuged for 5 minutes at 1500 rpm using high speed. Afterward, cell sediments were isolated, and the degree of apoptosis was measured using flow cytometry, followed by graph analysis.
3-1- Results of Chemical and Physical Properties of the Fusion Peptide
The R-type Pyocin, Enterocin A, and Lactocin genes were initially sequenced using NCBI website in order to determine their existence. Next, the recombinant sequence was complemented with an anti-cancer sequence. In the recombinant sequence, the specific ligand for the AGS cell line was designed using the BDB software. With IACP software, we determined whether the recombinant sequence is anti-cancer.
The Gene Script online server confirmed protein expression at each step. Among the practical and important factors is the Codon Adaptation Index (CAI). Using the JCAT web tool, it was optimized for the E.coli bacteria. We calculated the GC percentage and codon compatibility index. The CAIs were 1 for GC and 55% for GC expression, respectively.
A program called "mfold" determined the structure of the mRNA. According to the results, despite the lack of translation efficiency in the new host, mRNA stability was sufficient for efficient translation. The recombinant protein secondary structure (formed by RNA) minimum free energy was ΔG=-212.50 kcal/mol.
The profiles of recombinant proteins were extracted using ExPASy (Expert Protein Analysis Software). Based on the results, the following conclusions can be drawn. The molecular weight was 25823.48 Daltons, and the isoelectric point was 8.65 for the recombinant protein. Also, there were 236 amino acids in the protein. In the recombinant protein, 25 amino acids were negatively charged, and 29 were positively charged.Based on the predicted half-life of this protein, it is anticipated to last more than 30 hours in mammalian reticulocytes (in vitro), over 20 hours in yeast (in vivo), and over 10 hours in E.coli (in vivo). It appears that the protein has a stability index of 28.92 on ProtParam, which indicates that it is classified as a stable protein. This study is based on the mean grand-average hydropathicity index (GRAVY) of -0.31 and the aliphatic index (AI) of 80.25.
3-2- Result of Secondary and Tertiary Structure Analysis for Fusion Peptide
GOR4 was then used to review the structure of the analysis. Among the predicted secondary structures of the recombinant protein, 34.75% are alpha helices, 47.88% are random coils, and 17.37% are extended strands.
The three-dimensional structure, configuration, and stability of recombinant sequences were assessed with I-TASSER. A protein's secondary structure can be predicted based on its sequence using this software, including its alpha helix, beta strand, and random coil. As well, it predicts the normalized B-factor, which indicates the extent to which residues in proteins are thermally mobile. It is assumed that negative values indicate a relatively stable structure of the amino acids under experimental conditions. Additionally, this software predicts the three-dimensional structure of recombinant proteins. Among the potential factors that could have a significant role in the development of the proposed three-dimensional models are those such as the C-score, TM-score, and RMSD.
3-3- Result of Column Chromatography Purification
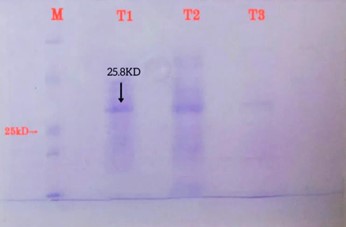
1 ml of eluted fractions was collected, and the fractions that contained the target protein were evaluated by SDS-PAGE. The result of SDS-PAGE on the product of ultrasound and protein purification and the purified recombinant protein showed that it weighs about 25.8 kDa (Figure 1).
3-4- Evaluation of recombinant protein expression using western blotting
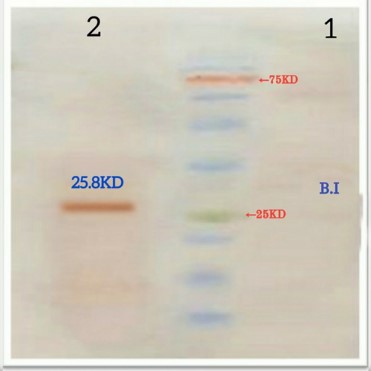
The recombinant protein with a histidine sequence and molecular weight of 25.8 KD was confirmed as specific by western blotting with a monoclonal anti-his tag antibody (Figure 2). In total, the concentration of the purified protein was 1000 μg/mL.
3-5- MTT Test Results
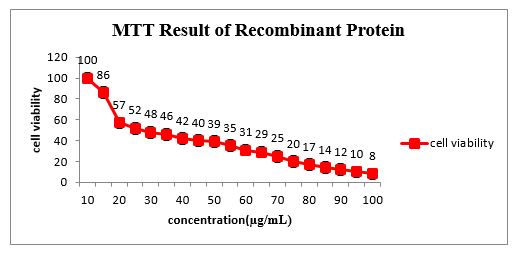
Approximately 80% of AGS cells exposed to an 80 µg/ml concentration of the recombinant protein for 24 hours were destroyed (Figure 3). The concentration of fusion proteins was set at 80 µg/mL.
|
|
|
|
Figure 1. Recombinant protein purification using nickel column (SDS gel). M: protein marker, T1: recombinant protein purified as a single band with a weight of about 25.8 kDa, T2: protein separated from the column using separation buffer, T3: protein separated using a washing buffer. |
Figure 2. Recombinant protein Western blotting. NO.1: Pre-induction deposition (B.I), NO.2: Post-induction deposition 2 hours |
Figure 3. MTT test results
3-6- Experimental results of flow cytometry in investigating apoptosis
As apoptosis began, phosphatidylserine (PS) was transferred to the outside of the cell membrane. Calcium enhances the binding of Annexin V to PS. In these conditions, fluorescein isothiocyanate was conjugated to Annexin V. Staining was done using a Propidium iodide solution (PI). The attachment of a compound to DNA occurs only when the plasma membrane structure of the cell has been destroyed. Consequently, cells passing through the first, middle, and late stages of apoptosis are distinguished from those that are passing through the last one. In addition, the frequency of cancer cell death is determined by necrosis, as well as early and late apoptosis.
The flow cytometry results are presented in Figure 4 and are as follows:
-
The Annexin V- /PI- region contains living cells.
-
The Annexin V+/PI- region was where the primary apoptotic cells were.
-
There was secondary apoptosis and necrosis occurring in the Annexin V+ / PI+ region, indicating the location of dead cells in this region.
-
An area containing Annexin V-/PI+ antibodies was damaged during preparation (necrosis with no apoptosis).

Figure 4. Two-parameter histogram. Section A: untreated cells. Section B: treated cells with recombinant protein
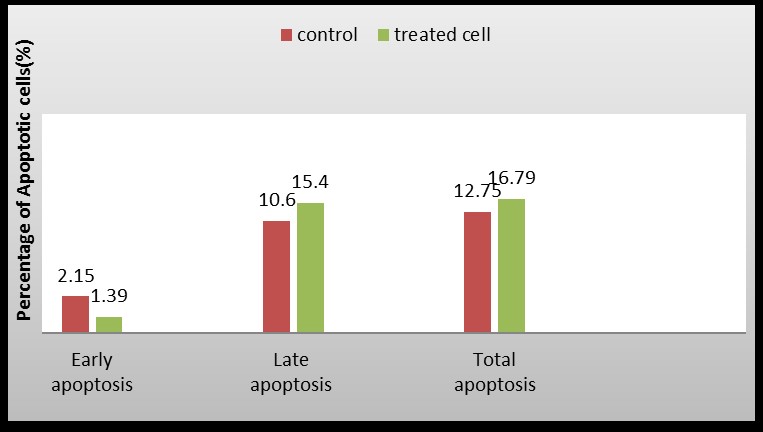
In comparison with nontreated cells, recombinant protein-treated cells had an increase in early apoptosis (Q3 region), late apoptosis (Q4 region), and necrosis (Q2 region), as shown in Figure 5. There was a significant difference between the untreated and treated groups in the number of healthy cells (Q4 region). According to these results, recombinant protein added to the medium at a concentration of 80µg/mL induces apoptosis in gastric cancer cells.
Figure 5. Comparison of apoptosis rates in treated and untreated cells by recombinant protein.
The primary objective of this research was to investigate if fusion protein, R-type Pyocin, enterocin A, Lactocin, and specific ligands possess an apoptotic effect on a gastric cancer cell line (AGS). An analysis of the anticancer effects of peptide fusion, bacteriocins, and specific ligands on the AGS cell line was conducted in our study. Generally, the recombinant protein induced apoptosis in the AGS cell line.
Recombinant protein-treated cancer cells had a significantly higher rate of apoptosis than untreated cancer cells, based on flow cytometry results. This confirms that treatment of the AGS cell line with recombinant protein at 80 μg/mL concentration induces apoptosis.
In many situations, this recombinant protein may be useful. For example, it can be used as an adjuvant for chemotherapeutic drugs or as a means of controlling apoptosis-induced genes affecting cancer cell proliferation and growth. Using 80 µg/mlL of recombinant Enterocin A, Pyocin, and R-Lactocin, this study demonstrated that 50% of treated cells with fusion proteins were killed after 24 hours.
The pET22b vector with the N-terminal pelB secretion signal was used in the present study. This signal sequence can ideally be used as a strategy for directing bacteriocin expression at E.coli periplasm. Protein secretion is directed by the leading pelB sequence in the pET-22b vector to the bacterial periplasm which may inhibit the growth of inclusion bodies in the cytosol, which results in the expression of soluble proteins in higher proportions (13).
In addition to diagnosis, the histidine sequence is used as a tag for facilitating recombinant protein purification and detection. When considering the advantages and disadvantages of the histidine affinity tag and Immobilized Metal Affinity Chromatography (IMAC), it is necessary to consider the histidine affinity tag's numerous advantages. It is easy to add this tag to the protein of interest, and, when purifying highly expressed proteins through IMAC, it easily achieves purities of up to 95% and 90% recovery of the tagged protein. In most cases, the histidine affinity tag does not interfere with protein function because of its relatively small size and charge. IMAC resin can elute histidine-tagged proteins under mild conditions, which enables them to remain biologically active. In addition, IMAC is an inexpensive and rapid method for purifying tagged proteins compared to other affinity protein purification techniques. Furthermore, polyhistidine affinity tags are commonly incorporated into recombinant proteins' N and C termini. Tag placement needs to be protein-specific in order to be most effective.
A disadvantage of using histidine affinity tags is the nonspecific binding of untagged proteins. Some cellular proteins contain two or more adjacent histidine residues, despite histidine being relatively uncommon (2% of all residues in proteins are histidine). There is a possibility that these proteins will coelute with the protein of interest, leading to serious contamination of the final product. Other systems, such as E. coli. Mammalian systems, tend to exhibit this problem more pronounced. For instance, these systems have a higher rate of consecutive histidine residues in proteins. Contamination can also occur as a result of disulfide bond formation between the proteins of interest and other proteins. In general, this problem can be avoided by using buffers containing 10 mM 2-mercaptoethanol throughout the loading, washing, and elution processes. Copurification with the desired protein can also be caused by nonspecific hydrophobic interactions. It is possible to reduce the interactions without significantly impacting the binding of the tagged protein to the Ni2+-NTA matrix by adding a small amount of the nonionic detergent Triton X-100 or Tween 20 to the protein buffers. Adding salt (up to 500 mM), ethanol (up to 20%), or glycerol (up to 20%) can also reduce nonspecific hydrophobic protein interactions in these matrices. Proteins should be tested experimentally to determine optimal levels of these buffer components (14).
Likewise, AGS cells exhibited the greatest level of apoptosis after protein fusion. Nisin-enterocin-epidermicin protein fusions increased apoptosis in AGS cells. As an alternative treatment, antimicrobial peptides, including bacteriocins, are gaining attention. It also seems that the fusion protein can be studied further and may be a future treatment for stomach cancer (15).
Furthermore, Pirkhezranian et al., found that Enterocin P expressed within pcDNA3.1 vectors inhibited the growth of mouse embryo fibroblast cells, liver cancer cells, colon cancer cells, and muscle skin melanoma cells. C26 and SW1353 as cancer lines were inhibited successfully by Enterocin P with an IC50 equal to 0.32 mg/mL and with an IC50 equal to 1.36 mg/ml. Researchers found that bacteriocins can affect specific cell lines (16).
Enterococcus Mundtii strain C4L10 was studied for its anticancer effects on oral, breast, and colon cancerous cells in research led by Yusuf et al., This bacteriocin has been identified as having an antiproliferative mechanism. It exerts its anticancer effects on the cell lines under investigation (17).
An earlier study by Paiva et al., determined when the cells were treated with nisin, the resulting IC50 values were 105.46 and 112.25 µM for breast cancer and liver cancer cell lines, respectively. In contrast to this research, the recombinant peptide of Enterocin A, R-type Pyocin, and Lactocin used in this study was far more effective against cancer, which could be because of the synergistic activity of these two bacteriocins (18).
There are several types of proteins in the Bcl-2 family that contribute to the regulation of apoptosis, such as the human Bcl-2 family of proteins that were studied in this study. Moreover, it prevents caspase-3 activity in the internal apoptosis pathway. In the apoptosis regulatory protein cells, the factors that may cause apoptosis are precisely balanced with those that suppress it. In terms of determining the survival or apoptotic death of cells, the Bax/Bcl-2 ratio is most strongly associated (19). Researchers found that recombinant Enterocin A, R-type Pyocin, and Lactocin increased Bax expression while simultaneously decreasing Bcl-2 expression (10). An indicator of apoptosis activation is the ratio of increased Bax gene expression to increased Bcl-2 gene expression. Using the bacteriocin Nisin against colon cancer cell lines, Ahmadi et al., demonstrated a balance between pro- and anti-apoptotic factors. Using Nisin at different concentrations significantly increased the ratio Bax/Bcl-2, demonstrating that cancer cells were drawn toward apoptosis (20).
In our study, a recombinant peptide consisting of Enterocin A, R-type Pyocin, and Lactocin increased caspase-3 expression and thus activated apoptosis via the intrinsic pathway. Ankaiah et al., discovered that enterocin A has anti-cancer activity against HeLa cells, gastric cancer, colon cancer, and other tumors. Fluorescent microscope observations include morphological changes of markers of apoptosis, including membrane depolarization, apoptotic bodies, and nuclear fragmentation (21).
Another study by Ankaiah et al., found that enterocin-B, a bacteriocin produced by E. faecium, has anticancer properties against HeLa, HT-29, and AGS cancer cells and that heterodimer enterocin-A+B significantly increased anticancer activity (22).
Apoptosis can be detected in treated cells using Annexin V-FITC and PI dyes. As a part of this study, using this technique, we investigated the effects of an 80 µg/mL concentration and a time of 24 hours of recombinant fusion peptide as tested on gastric cancer cells. We found that around 16.79% of the treated cells exhibited signs of apoptosis. 1.39% had early apoptosis, and 15.4% had late or delayed apoptosis. In comparison to the control group that did not receive any treatment, the treated group was found to have a lower percentage of viable cells as well as a higher percentage of apoptotic cells. Based on these results, we can conclude that our desired peptide causes apoptosis in a cancer cell line. In similar research, the induction of apoptosis in HeLa and HT-29 was observed using the flow cytometric assay of Annexin V-FITC / PI. Therefore, 53.93% of the cells exhibited delayed apoptosis, and 5.8% exhibited early apoptosis when Enterocin A was used at a concentration of 120 µg/mL, which influenced the number of cells that exhibited early apoptosis. This concentration of enterocin A resulted in 51.97 percent delayed apoptosis and 15.6% early apoptosis in the colon cancer cell line (21).
It was found that colicin N at concentrations of 5-15 µM killed a high percentage of different lung cancer cell lines, as compared to a control group that didn't get it (23). Using the MTT method and flow cytometry, Chumchalova et al., examined four colicins, E1, E3, U, and A on eleven tumor cell lines with mutations in their P53 gene. Five tumor cell lines were tested using flow cytometry and it was found that three pore-forming colicins (E1, U, and A) caused cell cycle arrest. There is also evidence that Colicin E1 and A have the ability to induce apoptosis up to 7-58% in three cancer cell lines (24).
According to Nami et al., enterocin showed an anticancer property when used in four cancer cell lines (AGS, MCF-7, HT-29, and HeLa) as well as a normal cell line (HUVEC). In their study, Ent.faecalis metabolites also induced cancer cell apoptosis (25).
The cytotoxicity of Pyocin S2, a Pseudomonas aeruginosa product, was investigated by Abdi-Ali et al., using HepG2 and Im9 cancer cell lines as well as normal HFFF cells. The findings showed that Pyocin S2 inhibited tumor cell lines, whereas there was no inhibitory effect on the normal HFFF cell line (26). The cytotoxicity of Pyocin S2 against human malignant cells and normal human cells was also investigated in a similar study by Watanabe et al., who found that with increased Pyocin S2 activity, the inhibitory effects on cells also increased (27).
Using cytoplasmic fraction from Lactococcus lactis, Kim and colleagues studied anticancer properties. This research indicates that a cytoplasmic fraction of Lc Lactis inhibits cell proliferation in human colon cancer cells (28).
In a recent study conducted by Fathizadeh et al., it was discovered that bacteriocins including Colicin, Nisin, Pediocin, Pyocin, and Mirocin may inhibit tumor growth at various levels by interacting with different molecular signaling pathways to either activate or inhibit tumor growth (29). Additionally, they demonstrated that Enterocin A and Colicin E1 fusion peptide had an antibacterial effect and could be used to treat or prevent bacterial infections (30). As part of another study (2021), the researchers used a combination of two bacteriocins, colicin E1, and enterocin A, against the AGS cell line. Based on the analysis of flow cytometry, annexin V-FITC/propidium iodide results were also able to reveal that the treated cells were more susceptible to apoptosis than controls (29).
In conclusion, our results indicate that recombinant proteins can cause cytotoxicity in AGS cell lines, suggesting that this recombinant protein activates intrinsically apoptotic pathways, resulting in cell death. The following specific outcomes were obtained through the present study:
1. A number of tests have been conducted to determine whether the recombinant protein can inhibit cell proliferation and induce apoptosis in cancerous cell lines. They showed that 80 μg/mL concentrations of the recombinant protein could cause apoptosis and cell death in stomach cancer cells.
2. Generally, recombinant proteins can be used to treat microbial infections and cancer; however, to clarify the exact mechanisms of the function of recombinant proteins, additional research is necessary.
These results can serve as a foundation for further studies on the potential of this recombinant protein in cancer treatment. The properties of this protein are consistent with what is expected from in vivo studies based on its in vitro findings.
Adapted from Neda Jalalvand's Ph.D. thesis, this article fulfills the thesis requirements. It would be a pleasure to thank the Department of Microbiology and Applied Virology Research Center of Baqiyatallah University of Medical Sciences, Tehran, Iran for cooperating throughout the entire research process.
This study was approved by Islamic azad university- Kazerun branch ethics committee under IR.IAU.KAU.REC.1398.153. All methods were carried out in accordance with relevant guidelines and regulations.
None.
Conflicts of Interest
The authors declare no conflict of interest.
D.E., N. J., M. R., M, M., and S.N. designed the study. N.J. performed research. D.E. performed data analysis. D. E. and N.G. wrote the article. M.M., S. N., and M.R. read and approved the final manuscript.
Received: 2022/06/30 | Accepted: 2023/01/28 | ePublished: 2023/03/30
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |