BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1685-en.html

 , Mojtaba Sadeghimanesh1
, Mojtaba Sadeghimanesh1 
 , Abbas Morovvati1
, Abbas Morovvati1 
 , Mohammad Soleimani2
, Mohammad Soleimani2 

 , Rohollah Mirjani3
, Rohollah Mirjani3 
 , Seyyed Hossein Mousavi4
, Seyyed Hossein Mousavi4 

2- Department of Microbiology, Faculty of Medicine, AJA University of Medical Sciences, Tehran, Iran ,
3- Department of Genetics and Advanced Medical Technology, Faculty of Medicine, AJA University of Medical Sciences, Tehran, Iran
4- Department of Cardiology, School of Medicine, AJA University of Medical Sciences, Tehran, Iran
The bacterium Bartonella quintana is the cause of Trench fever (1). B. quintana is a rod-shaped, optional, intracellular, and gram-negative bacterium that belongs to the subgroup of proteobacteria (2). This bacterium has negative catalase and oxidase reactions. B. quintana can be grown in axonal and cell culture media (3, 4). Its primary carrier is also known as the human body lice (5, 6). Bacteria are transmitted by lice bites or the entry of a pathogen through scratches on skin infected with lice stools. Lice stools usually become infected 5-12 days after eating and are contagious for the rest of the lice's life (7). Human reservoir B. quintana (8) and human body lice are its common carriers (9). B. quintana is present in erythrocytes during asymptomatic bacteremia and has been observed in bone row erythroblasts in bacteremia patients (10).
This bacterium has tropism for endothelial cells that leads to angioproliferative lesions, resembling bacillary angiomatosis (11, 12). The 1.6 MB genome of B. quintana has recently been sequenced and it was found that derivatives of the larger genome of 1.9 MB Bartonella henselae are the main differences between the two species without genomic islands in B. quintana (13). Both B. quintana and B. henselae genomes are abbreviated versions of chromosome I of Brucella melitensis, a highly phylogenetically related bacterium (13). Comparison of the genomes of B. henselae and B. quintana and their specialization in the human reservoir and lice transporter shows that the use of host-restrictive vectors is associated with accelerated genome degradation (13).
Infection caused by this microorganism was first recorded in soldiers during World War I, but is now seen in Europe, Asia, and North Africa (14) and has a global spread (15). In the early twentieth century, B. quintana infection emerged as a major source of morbidity and mortality among soldiers and was commonly known as Trench fever. During World War I, more than one million soldiers contracted Trench fever, and physicians in the military reported the disease in a wide range (7). It is now recognized as an emerging pathogen among homeless populations in US and European cities and is responsible for many conditions, including chronic bacteremia, endocarditis, and bacilli angiomatosis (16). Contemporary B. quintana infections have disproportionately affected homeless people, especially those with chronic alcoholism (17).
Symptoms include fever, headache, restlessness, pain, and tenderness, especially in the leg area, and are not usually fatal. Clinical signs of the disease may recur for seals after the initial infection and possibly subclinically. In some cases, osteomyelitis and bacilli angiomatosis can occur in people with immune system problems (6).
Infection with this bacterium is usually seen in two ways. The first is an opportunistic and febrile infection commonly seen in AIDS patients called bacilli angiomatosis, most of whom have been exposed to head or body lice (18). The second is a febrile illness that is usually transmitted by lice. This type is more common in the homeless and addicts. Bartonella spp. It can cause acute or chronic infections in cats, dogs, and humans and has a variety of clinical manifestations such as intermittent fever, granulomatous inflammation that affects the heart, liver, lymph nodes, and other tissues, and endocarditis, polycystic angiosis, and bacilli. Vasoproliferative tumors have been reported in cats, dogs, and humans (19). Some researchers speculate that erythrocyte infection may allow the body lice to effectively transmit B. quintana (17). Although researchers have reported sporadic prevalence of contemporary B. quintana infections, serological studies have shown that pathogen exposure is more common in some high-risk populations than in overt clinical diseases (20).
Serological tests are the most widely used method for diagnosing Bartonella infection. Indirect immune-fluorescence is the reference method. In the indirect immunofluorescence method, the titer of immune-globulin G 21:50 indicates Bartonella infection, and the titer 21: 800 predicts endocarditis (21).
Bacteria are detected by blood culture in an agar medium, and colonies are usually seen after 8-21 days because they grow late (22, 23). Molecular detection of Bartonella species can be performed using PCR of blood and tissues. Various tissues may be used, including lymph nodes, heart valves, skin, and liver (16). Although fresh tissues are more comfortable, formalin-proven paraffin tissues may also be used for PCR-based assays. The use of universal primers for amplifying the 16S rRNA gene is not a suitable method for detecting Bartonella infection at the cheek level because the 16S rRNA genes are more than 97.8% similar to Bartonella species (16). In fact, the longtime of detection through culture and new targets for identifying Bartonella species has led to the importance of molecular methods such as PCR for more accurate and faster detection. In fact, the diagnosis of B. quintana infection is challenging and the most common methods used to diagnose acute and chronic infections are specialized microbiological culture methods, polymerase chain reaction (PCR), immunohistochemistry (IHC), and serology.
Definitive diagnosis of B. quintana infection is made by isolating the organism from a blood or tissue culture. However, because B. quintana is very difficult to isolate in culture, most diagnoses are based on supportive diagnostic tests, such as serology and PCR (20). In general, serological testing of B. quintana should not be used to diagnose B. quintana infection and should be interpreted in terms of epidemiological features and clinical manifestations (20). Therefore, in this study, the infection of B. quintana with molecular methods such as PCR was investigated.
DNA Extraction
At this stage, 100 samples of negatively cultured endocarditis were collected from selected military hospitals. DNA extraction from B. quintana was performed with the DNP extraction kit of Sinaclone Company (Iran, DNP; lot No. EX6071) and according to the kit instructions. The concentration and quality of the obtained DNA were measured using a nanodrop device.
Primer Design
For PCR, the primers were designed using Gene runner software (version 2) and CLC sequence viewer (version 6) and made by Sinaclon (Iran). In order to finalize the designed primers and ensure their specificity, the primers were blasted using the information registered on the NCBI website, and their accuracy was confirmed.
Reproduction and Isolation of B. quintana ftsz Gene
For each PCR, a 25 μL reaction mixture was prepared to contain 12.5 μL of the Master mixture, 1 μL of each primer (F2: 5'-CTCAACTTCAAAGACAGGCGA-3 'and R2: 5ˊ-TGACTTTTAGGGCGGAACTG-3') with a concentration of 10 ppm, 5 μL of template DNA and 5.5 μL of water were prepared by PCR. Two samples were prepared for the positive control (B. quintana; 172 bp) and negative (water equivalent was added instead of DNA).
PCR according to the method (24) under initial denaturation at 94°C for 3 minutes, followed by 35 denaturation cycles at 94°C for 30 seconds, annealing at 52°C for 35 seconds, and expansion at 35 cycles. 72°C for 35 seconds, with a final extension at 72°C for 5 minutes. Then electrophoresis was performed on 2% agarose gel, and the gel was stained with Red Safe (25).
Positive Control Synthesis
Due to the fact that a positive sample was not available for bacteria, positive control for these bacteria was synthesized based on the sequence of primers and prepared in PUC 18 vector.
CTCAACTTCAAAGACAGGCGACTTCAATTCGTAAAAATGATCCTGGAATGTCTCAGACTTCTTTTCATCTTCAGTCACCACCCTTGCGTTCTGAGTCAATGGTAGAAGTAATAGAAGCACTTGAAATAGAAAGGGCAA
Feature Determination
In order to evaluate the specificity of primers and determine the specificity of the method, PCR reaction with the conditions mentioned above was performed on the genome of negative control samples. Also, to ensure the ability of the extracted DNA to be used in the PCR reaction and the absence of inhibitors in it, the PCR reaction on the DNA of bacteria (Staphylococcus aureus (ATCC 25923), Bacillus subtilis (ATCC 6051), Shigella sonnei (ATCC 9290), Escherichia coli (ATCC 25922), Enterobacter faecalis (ATCC 29212) and Pseudomonas aeruginosa (ATCC 27853)) were used as positive controls to determine specificity.
Cloning of the B. quintana ftsz Gene
To clone part of the B. quintana ftsz amplified gene in plasmid PUC 18, first, the PCR product using the Bioneer gel extraction kit protocol (South Korea, Cat No. K3037 AccuPrep® PCR / Gel Purification Kit); Purified. Cloning was performed using SinaClon TA Cloning Kit (Iran, SinaClon TA Cloning Kit; Cat No. CL5841) according to the kit instructions and with PUC 18 vector. After ligation, JM107 E. coli susceptible to calcium chloride was used to accept the plasmid containing the gene (26). After susceptibility of the bacteria to plasmid acceptance, 10 μL of the plasmids containing the gene was added to the microtube containing the susceptible bacterium and finally transferred to the bacterium by heat shock using the recombinant plasmid. The bacteria were then placed in a sugar incubator at (150 rpm, 3h 37°C) for 60 minutes. Transformed bacteria were cultured on LB Broth medium containing Ampicillin antibiotic. The culture medium was then centrifuged at 9000 rpm for 2 minutes. From residual precipitate on plate surface (50ml) LB agar containing (50 mg/mL) Ampicillin 100L, 100L X-gal (20 mg/mL), 80L IPTG (24 mg/mL) and Tet (10 mg/mL) 100L , Added and broadcast. The plate was placed in a sugar incubator at 37°C for 16 hours. The following methods were used to confirm the cloning and transfer of recombinant plasmids into bacteria.
Formation of white Colonies Containing Recombinant Vectors
After culture, the transformed bacteria are seen in the culture medium of blue colonies (colony without recombinant plasmid) and white (colonies containing recombinant plasmid).
PCR Colony
For PCR colony, 2 to 3 white colonies from the transformation stage were selected, and PCR reaction was performed using PCR Master mix (Ampliqon; Cat. No: A190303). The product was then electrophoresed in 1% agarose gel to confirm the presence of the cloned fragment.
Extraction of Recombinant Plasmid
Plasmid extraction was performed after confirming the presence of recombinant and inserted plasmids using ViVantis Plasmid Extraction Kit (Malaysia, Plasmid DNA Extraction Kit; Cat No. GF-PL-050 GF-1). In a quantitative evaluation of plasmid extraction, the first 1:10 and 1:100 dilutions were obtained from the plasmid extraction sample. After reading the concentrations of the dilutions prepared by the nanodrop device, the final concentration of the extracted plasmid was obtained in a volume of 100 microliters. Sensitivity and Detection Limit
To determine the sensitivity and limit of detection (LOD), the minimum number of copies of the desired fragment that can create a clear band in the PCR reaction was calculated. For this purpose, double dilutions (1-10 to 10-10) of PUC18 plasmid with specific concentration were prepared, and PCR was performed using them, and its products were electrophoresed on 1% agarose gel, the last dilution in which the PCR product bonds, will be the LOD that can be obtained with the relation CN = MN / LD, the least number of traceable copies.
DNA Extraction
Pure DNA was obtained using a nanodrop device from samples of patients with endocarditis. By examining the result of quantitative DNA analysis, its amount was calculated between 1.69 and 1.8.

Figure 1. Extracted DNA sample (L: DNA Ladder and 5-1: Purified DNA)
Based on the collected samples, endocarditis was examined for the presence of B. quintana in these patients, but out of 60 samples collected, no positive samples were reported for any of these factors.
Specification
In order to investigate the specificity for each pair of primers, PCR reaction with the conditions related to each gene was performed on the genome of the negative control bacteria extracted. Still, no band was observed after electrophoresis of PCR products related to each gene on the agarose gel (Figure 3). This indicates that the designed primers attach only to the target bacterium.

Figure 2. B. quintana; L: DNA Ladder, 1: DNA, size: (172 bp)
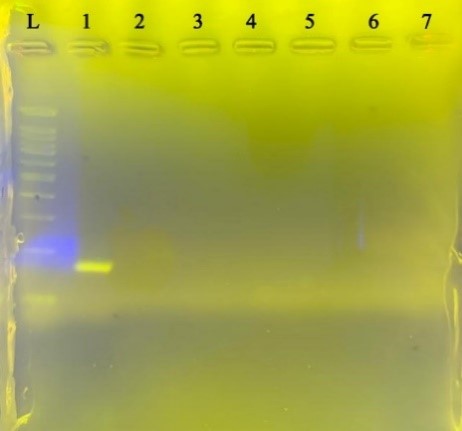
Figure 3. Specification for B. Quintana
Negative control bacteria from left to right L size marker, B. quintana, Staphylococcus aureus, Bacillus subtilis, Shigella sonnei, Escherichia coli, Enterobacter faecalis, Pseudomonas aeruginosa, Klebsiella pneumoniae, respectively
Determine LOD
The last dilution of PUC18 plasmid for B. quintana with an initial concentration of 780 ng/µL, which formed a detectable band on the gel, was calculated to be 7-10, and the minimum number of detectable copies in a 25 μL PCR reaction equal to 24 copies (Figure 4).

Figure 4. B. quintana detection limit.
Clinically, diagnostic interpretation of the results of serological and molecular tests that support Bartonella exposure or infection, respectively. But it is challenging in patients without normal clinical manifestations or environmental epidemiological exposure (27). For example, the diagnosis of B. henselae infection in a patient with lymphadenopathy after being bitten or scratched by a mammal (cat or dog) is simple, and microbiological tests are often unnecessary to confirm the clinical diagnosis (27). In contrast, syndromes or diseases caused by Bartonella spp. Such as endocarditis, pleurisy, angiomatosis, fever of unknown origin, isolated liver abscesses, etc. are clinically challenging and diagnostically problematic and require microbiological confirmation of the diagnosis.
In addition, the specific definition of Bartonella spp. As an etiological factor of the disease, it is necessary to determine the epidemiology of the disease and facilitate disease monitoring among different populations (humans, animals, and vectors) (28). However, implying a Bartonella spp. As the cause of an unusual clinical presentation when using a serological test can be difficult, even if the bacterial DNA is amplified from blood or enriched blood culture or if the organism is cultured from blood (29); Therefore, it is possible that Bartonella spp. Causes long-term intra-erythrocyte and endothelial cell infections in healthy individuals without long-term pathology or bacteremia. Or it may lead to chronic and slow-progressing vascular injury in an unsafe person (14, 30).
In addition, long-term bacteremia of B. quintana has been reported in the homeless and in immune-compromised patients (6). Regardless of the microbiological or molecular blood culture results of Bartonella in this study, patients without a clinically defined medical syndrome should be interpreted with caution.
In most clinical microbiology laboratories, the diagnosis of Bartonella infection is based on serological assays, with current techniques lacking sensitivity, specificity, or both, in addition, due to poor overall sensitivity in isolating Bartonella species from conventional cultures (gold standard). Not used. In recent years, molecular assays have been used for clinical diagnosis and epidemiological studies in vectors (such as fleas) (31, 32).
Due to their strict nature, Bartonella species are good candidates for species identification by PCR-based method. Many attempts have been made to develop rapid and specific molecular methods for detecting Bartonella because of the known benefits over culture (33). PCR seems to be a useful tool that may reduce the detection time of B. quintana during systemic infections, especially chronic bacteremia (34).
PCR itself is a sensitive method for detecting various pathogens (35, 36). DNA extraction methods are not suitable for processing large amounts of blood. For this reason, it seems to be the optimal example for diagnosing Bartonella infection in humans by PCR, lymph node, or tissue biopsy (including heart valves in cases of endocarditis in case of surgery) or aspiration. B. henselae DNA has been consistently described in cat blood samples, reflecting long-term bacteremia in cats. In humans, the detection of Bartonella DNA in patients's blood has been described as sporadic (37). Researchers have confirmed that in a small number of cases, it is possible to detect Bartonella DNA in serum (38). This may reflect temporary bacteremia or other factors. The study of Vermeulen et al. (2008) showed low sensitivity (18%) of PCR method in the diagnosis of Bartonella (37). The results of this study show that PCR using serum has a limited value in the usual clinical microbiology laboratory. Accordingly, PCR may be more valuable on plasma or whole blood samples. The moment of sampling is also very important in diagnosing Bartonella. It is recommended that these be used in future studies to better diagnose Bartonella.
Due to the fact that this negative culture of endocarditis is more common among the homeless or people prone to lice, it seems that a wide range of different people will be considered in future studies. To be able to confidently present management strategies in the field of prevention and treatment. In addition to blood samples, lymph nodes and tissues are also recommended in studies. In addition to different examples, evaluation of different PCR methods as well as the use of serological tests along with PCR can also increase the accuracy and reliability of the diagnosis.
Conflicts of Interest
The authors declare any conflict of interest.
Received: 2022/02/27 | Accepted: 2022/05/13 | ePublished: 2022/08/8
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |






