BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1435-en.html
2- Department of Microbiology, School of Medicine and Applied Microbiology Research Center, Institute of Toxicology and Systems Biology, Baqiyatullah University of Medical Sciences, Tehran, Iran
3- Department of Microbiology, School of Medicine and Clinical Research Development Committee, Baqiyatullah University of Medical Sciences, Tehran, Iran ,
Pseudomonas aeruginosa (P. aeruginosa) a bacterium which is rapidly resistant to antibiotics due to chromosomal changes and genetic mutations caused by plasmids and transposons (1, 2) with the emergence and increase of bacteria survival. In the last two decades, drug-resistant infections could increase mortality each year, especially in burn patients and intensive care unit ward (3). P. aeruginosa has several causative factors, including biofilm formation (4), mucoid phenotype development (5) and type III secretory system. Controlling colonization potential in various parts of the body, including the formation of liver and lung fibrosis, the formation of dental plaque and tooth decay, plays a role in secondary infections of burns (6) are considerd. Moreover, the existence of diffusion pump systems and β-lactamases helps in the resistance of P. aeruginosa to penicillins, cephalosporins, carbapenem, monobacatom, fluoroquinolone, aminoglycoside and cholestine (7). The likelihood of resistance to these antibiotics increases due to mutations and the transfer of horizontal genes through plasmids, protons and transposons (8). The bacterium has a wide range of pathogenic factors, including exotoxins, lipopolysaccharides, pili, phospholipases, elastases, proteases, and other agents (9). Among the agents and virulence of this bacterium are exoA, exoS, exoT, exoU and exoY cytotoxins, which enter the target cells in the host through the type III secretory system (10).
However, the emergence of recent antibiotic resistance has created an urgent need to develop new waves of plant antibacterial agents or make them more effective than a combination of herbs and antibiotics to circumvent this problem. Medicinal plants produce active biological compounds such as phenolic acids, vitamins, flavonoids, and aromatic compounds such as terpenoids, steroids, alkaloids, and organic cyanides (11). In this regards, the plants are valuable sources of plants secondary metabolites that are used as medicines (12).
Ammi visnaga (A. visnaga) is widely found in North Africa, the Middle East, North America, Argentina, Chile, Egypt, India, Iran, Mexico, Tunisia, and Russia, and in Northern Algeria, and the extracts and periano quamarines include visnadin, samidin, and dihydrosimidine, have major clinical applications in angiography as well as diseases linked to asthma, atherosclerosis, coronary artery, circulatory problems, painful menstruation, skin treatment, and psoriasis (11). The ethanolic extract of the plant has shown antibacterial activity, which prevents the growth of Mycobacterium tuberculosis or any of its symptom even at very low concentrations. The effect of ethanolic extract of these plant against the Enterococcus faecalis is significant effect at high concentrations. Some other studies have shown the effectiveness of essential oil of this plant against Eucalyptus, Staphylococcus SPP, and P. aeruginosa (12). According to a recent study, the growth spurt of the MIC test for P. aeruginosa, Staphylococcus aureus, and Acinetobacter baumanni under the influence of ethanolic extract of dental plant was 25, 25, 25, and 50 mg/ml, respectively (13). A semi-quantitative RT-PCR method was applied to know the inhibitory effect of S. khuzestaniea plant on the expression of exoenzyme S, exotoxin A, and secretory systems of Pseudomonas aeruginosa has reported. Therefore, considering its clinical importance, further studies in this field are necessary to probability inhibition of pathogenic patterns and antibiotic resistance index genes in this bacterium. The purpose of current study was to investigate the plant (A. visnaga) extract on the expression of pathogenic genes: exoA and exoS which they are involve the antibiotic resistance and pathogenicity of P. aeruginosa.
Locality Address of the A. visnaga
The source of the plant “A. visnaga” used in this study is Iran, Khouzestan province; between Bagh- Malek and Izeh, 620m, Mozaffarian (43170, Research Institue of Forests and Rangelands- Tehran (TARI) herbarium, and identification by S.M.M. Hamdi.
Collection and Extract Preparation of A. visnaga
In this study, A. visnaga was collected from the Khouzestan Province Iran. This medicinal plant is native to that area of Iran. After separating the waste, the plant's aerial parts were first air-dried and then dried in the shade completely. In order to extract the samples, the plant materials dried by electric mill (having a sieve with holes of 2mm diameter) and were crushed and kept in glass containers and finally, the plant extract was prepared (Figure 1) using the Soxhlet method with some modification (14). In summary a sample of 16.6 grams of powdered dry leaves of the plant (Fig 1; part B) was placed inside the cartridge chamber. Then, 250 ml of ethanol was added inside the rounded-bottomed flask, and then the soxhlet was installed on the flask, and in order in order to maintain temperature the water to flow was used by open the tap and turn on the 60°C heater, and after ten minutes the ethanol temperature boiling was reached, the temperature was reduced and after 3 hours from the beginning of the extraction process (Fig 1; part C and D), the extraction process was finished. At this time, the heat was turned off and the system was allowed to cool down and the vapors were cooled in the refrigerant overnight. In order to removing the exceed remained ethanol from the liquid phase, the flask was subjected to a rotary device. Then, the contents of the flask were subjected to Lyophilized (frozen at -20°C and dried at -50°C under vacuum condition) and kept it on refrigerator until used.

Figure 1. A) The aerial part of the A. visnaga plant; B) Dried air part of the A. visnaga; C and D) Soxilet system for extracting aqueous extract of the A. visnaga plant; With thanks to Ark Laboratory for providing the Soxilet system for this Study.
Bacterial Strains and Storage Methods
P. aeruginosa used strain ATCC 27859, which producer exotoxin A and S were used. The bacteria were stored in the freezer at -80○C for one year. P. aeruginosa containing vial was removed from the freezer and its contents were gently melted. The sample was sub-cultured on fresh Brain Heart Infusion Agar (BHI Agar) medium and then placed in an incubator for 18-24 hours at 37 º C.
Cultivation of P. aeruginosa for Exotoxins Production
The bacterium was inoculated into BHI Agar culture medium and placed in an incubator at 37º C for 24 hours. After that, with a sterile loop a single colony has injected into a tube containing Brain Heart Infusion Broth culture medium. The initial optical density (OD) rate was taken out with a spectrophotometer periodically until growth rate obtained. The 4ml of a per culture was taken into an Erlenmeyer flask containing 100ml of Brain Heart Broth. The flask was placed in an incubator at 37○C, and the OD was measured with a spectrophotometer every hour. The dilution and the counting of colonies were also performed every 1 hour. But after reaching the logarithmic phase, the dilution and the counting of colonies were performed every 30 minutes.
MIC Determination of the A. visnaga Extract
In order to determine the minimum inhibitory concentration (MIC), the liquid dilution method or serial micro dilution based on the CLSI standard was used (15, 16). First, 1grams of the Lyophilized extract was dissolved in onemililiter (1 ml) of normal saline and vortexed, then the solution was sterilized with a syringe 0.45µm Millipore filter and finally the working extract solution was prepared. To determine the MIC, the micro dilution method was used in 96 well plates; hence, 300μl of BHI broth culture medium (Merck Germany) was poured into wells 1 to 9 in each row and then 100ul of the extract was added in all wells of the first column (by repeated triple). After that, the first well of the first row was mixed with the sampler. Then, 200μl of it was transferred to the next well which contain 200 μl of BHI broth with a sampler. This process was done till well 9, and 200μl was poured away from all of them. The extract concentrations in the wells were 25, 12.5, 6.25, 3.125, 1.562, 0.751, 0.39, 0.195, and 0/097 micrograms per well, respectively. Finally, 50 microliters of McFarland's 0.5 microbial suspensions were added to all wells and the results were read after 24 hours of incubation at 37○C. The last portion of figure 2, in which, no growth was considered as MIC, and almost kill 99.9 percent of bacteria was also considered as MBC.
RNA Extraction, Primer, and Real-Time- RT PCR
To evaluate the expression of the target genes, the containers and tubes used were free of RNA.
Using the extract RNA kit (RNA-protect Bacteria Reagent, kit; Cat No: 76506 and Proteinase K Cat no: 19131; Qiagen), the RNA was extracted based on the kit direction. The RNA extraction steps were performed in an environment free of any contamination and at the temperature of 25°C. After extracting RNA, its quantity and quality were evaluated using the Nano drop device and Agarose gel electrophoresis. The concentration and purity of the RNA sample were investigated using the Ultra Violet absorption at nm260 and 280 wavelengths. Also, the Kit direction cDNA synthesis (QuantiTect Reverse Transcription kit; Cat no: 205311 and quantiTect SYBER Green RT- PCR kit; Cat no: 204243. Qiagen) was used to make cDNA from extracted RNA, after determining the amount and concentration of the resulting RNA and normalizing the concentration. The selected primers were synthesized by Macrogen (South Korea). Table 1 shows the sequence of corresponding primers. Finally, in order to perform the Real- time PCR efficiently with SYBR Premix, a 35-cycle was considered as the final thermal cycle for the reaction. After the reaction, the products were subjected to 1.5% Agarose gel electrophoresis. In addition to gel electrophoresis, data diagrams were analyzed using Graph Pad Software Inc., San Diego (GRAPHPAD PRISM 6CA, USA). All preparation steps of the RT- real-time PCR mixture, the preparation method, and protocol were performed under aseptic conditions and sterile instruments.
| Gene Name | Sequence | Molecular weight | Reference |
| exoS F | CTACACCGGCATTCACTAC | 140bp |
(17) |
| exoS R | AAGTCTTCACTACCTGTTCAG | ||
| exoA F | CACGAGAGCAACGAGATG | 165 bp |
|
| exoA R | GGCGAGGTAGTTGTAGAC | ||
| 16s F | ATCACCTTCTACTTCG | 187bp |
|
| 16s R | CCAGAGCCATGTTGTACT |
Based on instructions, the working concentration was 0.8μL of the 10µmole of the forward primer and 0.8μL of the 10µmole concentration of the reverse primer were mixed for each marker.
MIC and MBC Determination
This study investigated the results of the minimum inhibitory as well as the minimum bactericidal concentration of A. visnaga extract (were 25, 12.5, 6.25, 3.125, 1.562, 0.751, 0.39, 0.195, and 0.097 micrograms per well). The lowest concentration with no turbidity was considered as the minimum inhibitory concentration, which were 3.125µg/well. The MBC test results were also presented, showing no bacterial growth at 3.125, 6.25, 12.5, 25 µg/well concentrations (Figure 2).
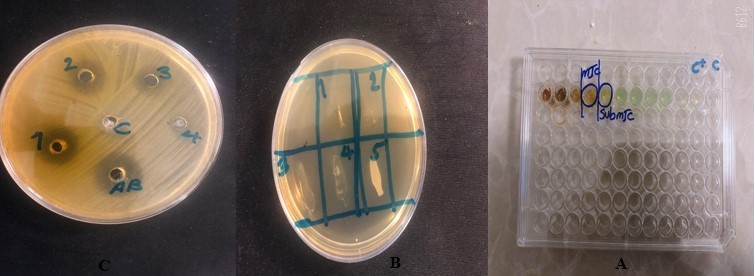
Figure 2. The results of Anti-pseudomonas effects for MIC and MBC
Preparation of Melting Curve
To perform the Real-time PCR reaction, the corresponding genes were designed, synthesized, and repeated by PCR twice. At the end of the reaction, a standard linear curve based on Ct and logarithm of DNA concentration was drawn to confirm the proliferation of the amplification product.
Electrophoresis PCR Product
The electrophoresis of the PCR product shows the overall PCR response along with the gene marker (exoA = 156bp. exoS = 140bp).
Real-Time- RT PCR Results
An analysis of the RT Real- time PCR results is presented in Figure 3, which shows the effect of the MIC concentration of A. visnaga extract on the expression of exoA and exoS genes of P. aeruginosa.
As shown in the above figure 3, the exoA and exoS genes in the control group are at the highest level. While the aforementioned genes are reduced after treating with the MIC concentration of A. visnaga extract, their expression levels decreased significantly, which is a sign of the positive effect of the A. visnaga extract on the reduction of Pseudomonas bacteria exoA and exoS toxins.
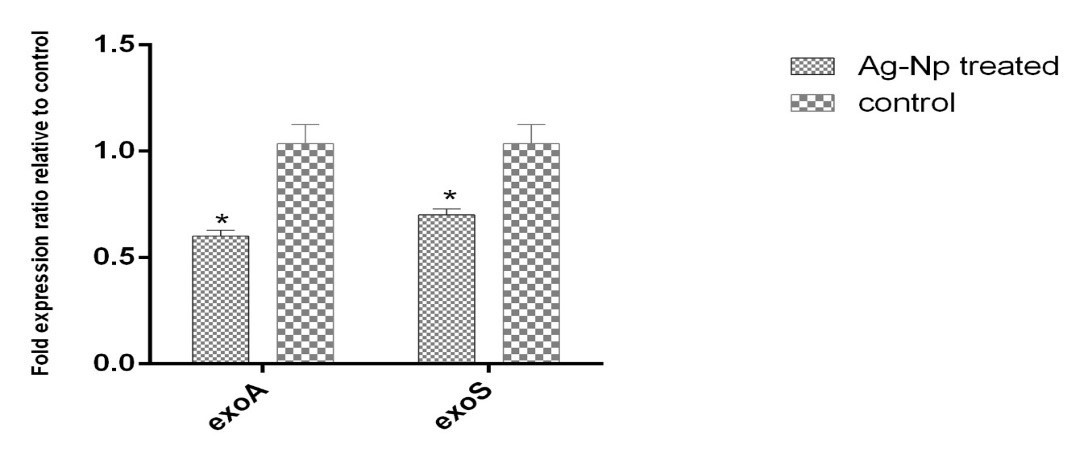
Figure 3. The diagram showing the amount of exoA and exoS genes expression ratio by using the Graph Pad Software Inc., San Diego (GRAPHPAD PRISM 6CA, USA) instrument for analyzing and figure out of the results.
Due to the widespread resistance of P. aeruginosa to antibiotics as well as antibacterial agents (18), led we to assay on the new substances by acting on virulence and pathogenesis genes has been considered. In this regards, exotoxin A and S of P. aeruginosa as virulence genes have isolated from human and animal samples are reported (19). This gene is secreted by the general secretory pathway and may altered therapeutic strategies (20). A research results reveal that, P. aeruginosa exotoxin A has regulated factors to corneal infection by cytotoxic and invasive in a murine scarification model (21). Another research results indicated that, Pseudomonas aeruginosa exotoxins injection into host cells can be modulated by host factors at the level of translocation assembly and/or activity and this phenomenon may involve to bacterium colonization and pathophysiology (22). A recent study has also identified that, ExoS as a double toxin - Rho GAP and ADP-ribosylating- activity (23).
This study intended to investigate the effect of A. visnaga aqueous extract on the expression of exotoxin A and S genes of P. aeruginosa. The results showed that the MIC concentration extract of A. visnaga reduced the expression of these genes, which are evolved on the pathogenicity of bacteria. The MIC test results found the lowest bactericidal concentration of A. visnaga extract as 3.125µg/well. A wider discussion on the subject and comparison of the findings seemed difficult given a scant previous researches in this field. However, some previous researches in the area could yield semi close and less similar antibacterial actions (24) Because the extraction solutions were different in the two studies. In our study, the aqous extract was used, while, the Rosina Khans has utilised Petroleum ether extract. This may causes diferensis related to detrimental effect in the active substance.
In this regards. the antibacterial effects of the present study were similar to those who concluded that the A. visnaga ethanolic extract inhibiting the growth of S. aureus, E. coli, K. pneumoniae and Candida albicans, was consistent with MIC 12.5 (25). Interestingly, while other researchers showed the effectiveness of A. visnaga against gram-positive and gram-negative bacteria, mechanisms of action had not been taken into account until now. In the other words, the present study is the first to report the expression inhibitory effect of the A. visnaga extraction on the pathogenic genes (exotoxin A and s gene). The potential of gene expression inhibitory effects has discussion as antioxidant as well as anticancer activity of the A. visnaga extracts (26) which reported by Aydoğmuş-Öztürk. Furthermore, Quorum Sensing Genes expression study in Pseudomonas aeruginosa has reported (27).
In a recent research, two main compounds of furanochromones of khellin (A. visnaga) have been described as potential herbicidal activity (28), but their effects on prokaryotic cells are not reported yet.
However, essential oils and aqueous extracts of A. visnaga which may penetrate the cell wall and cytoplasmic membrane of bacteria, making them particularly vulnerable to damage lipids, proteins, and DNA needs further studies in future.
In addition, some phenolic compounds in essential oils are oxidized due to their contact with ROS (Reactive oxygen species) and produce highly active phenoxy radicals (29).
The P. aeruginosa infection is very rare in healthy individuals, but as an opportunistic bacterium, it infects patients with immunodeficiency especially, cystic fibrosis, burns, or AIDS patients (30). Due to inherent resistance to various antibiotics or chemotherapeutic agents, P. aeruginosa, it is difficult to eradicate and therefore has a high mortality rate, because of exotoxins and antibiotic resistance related to toxins production (31, 32). However, the expression of exotoxin A and S genes in P. aeruginosa is very common; studies in gens expression inhibition are scant and a few conducted. So far, in this study we have only examined the effect of plant extracts on the expression of exotoxin A and S genes.
However, more study is necessitating, the results of the current research revealed a significant reduction on exotoxin A and S gene expression, it may have decline the colonization, pathogenicity and aggression. Such, the study was designed and conducted under this hypothesis.
The results of this study showed that the extract reduces the expression of pathogenic genes (exotoxins A and S) of P. aeruginosa.
The MIC test results also show the lowest 12.5 µg of the extract has bactericidal activity. It is really hard to compare and review specific sources since very little research has been conducted in this field. However, the overall results of the current study show that A. visnaga extracts can be reduced the causative and pathogenic factors of P. aeruginosa. However, in this study antibacterial effect of the A. visnaga extracts has shown, the gene expression inhibitory on exotoxin A and S were significantly. However, further research to determine the HPLC active gradient analysis are necessary. Although there was no possibility of HPLC analysis in this study and this is an important weakness of this study, but the effect of gene expression inhibition of the A. visnaga extracts was significant and may be effect on the pathogenicity of bacteria.
The authors would like to thank the Deputy of Clinical Development Medical Center of Baqiyatallah Hospital for their support and for their technical support. Also the authors would like to thanks to Ark Laboratory for providing the Soxilet system for this Study.
This study was supported by Vice-Chancellor for Research and Technology of Kermanshah University.
Conflicts of Interest
The authors declare no conflict of interests.
Received: 2022/05/28 | Accepted: 2023/02/2 | ePublished: 2023/03/30
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |








