BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1068-en.html
2- Department of Microbiology, Faculty of Science, Kerman Branch, Islamic Azad University, Kerman, Iran ,
.
Blood is one of the body's fluids that must be free of any microorganisms in a healthy state. However, under certain conditions, it may change and microorganisms may be transferred to the circulatory system and cause a blood infection. Currently, septicemia is one of the leading causes of death in critically ill patients in the intensive care unit (1,26). Gram-negative bacteria are among the most important causes of toxic septicemia, which has been on the rise in the last 20 years (2). The severity of the disease to some extent determines the outcome of disease. In fact, mortality might occur in septicemia in up to 30%, severe septicemia in 50% and septic shock in 80% (3). At least part of the mortality in septicemia is due to a lack of specific warning clinical signs in its early diagnosis. In addition, the lack of a specific infection marker to diagnose the disease has led to problems in distinguishing infectious agents from non-infectious ones. For this reason, many studies have been conducted in recent decades to access markers that can be used to detect early toxic septicemia. Some recent studies have reported an increase in serum pro-calcitonin levels in this group of patients (4). Cytokines play a major role in causing septicemia (5). The mechanism of induction of cytokine production in patients with septicemia has not yet been established. The gene expression of NLRP1 and NLRP3 inflamasomes following septicemia is one of the causes of inflammation and induction of cytokine production in this group of patients. Inflamasomes are a group of cytoplasmic receptors as multi-protein complexes of the innate immune system that are activated in parenchymal and non-parenchymal liver cells and include AIM2, NLRP3, NLRP1 and NLRC4 (6). Stimulation of each of these molecules activates Caspase 1 by the ASC molecule, followed by Caspase 1 leading to activation of Pro-IL-1 and Pro-IL-18 inflammatory cytokines (7). Some studies have suggested that NLRPs may be important regulators of inflammation in microbial infections. The present study was performed to compare the expression of NLRP1 gene expression in patients with toxic bacterial septicemia. Attempts are being made to use the Real time PCR method as a new assay in diagnosing comparisons between NLRP1 gene expression in patients with septicemia and the control group with a general understanding of the human immune system, especially inflammatory cytokines and the NLRP1 gene rely on the background of the research to be presented.
This cross-descriptive study was performed in 2018 during 6 months on 40 blood samples of patients with septicemia and 40 blood samples from healthy control individuals in Afzalipour Hospital in Kerman, as an available sample. Sampling was performed based on the hospital's routine treatment of patients and no blood sampling was performed solely for the present study. In addition, these patients were in the middle age group and did not have underlying diseases such as diabetes. 5 to 10 mL of blood from people with symptoms of septicemia was confirmed on a liquid blood culture medium with the approval of a specialist doctor, and blood culture glasses were incubated for 48 hours at 37°C (8). Then, blood culture was performed in aseptic conditions on Blood Agar and Mc-Conkey agar culture medium by streak method. From the grown and positive samples, bacteriological diagnostic tests including Gram staining, biochemical and diagnostic tests were performed to identify the bacterium causing infection (8).
RNA Extraction and cDNA Synthesis
Buffy coat from a blood sample, 40 patients with septicemia and 40 healthy controls were obtained. The RNA extraction was performed using the based RNX-Plus kit (Cinaclon, Iran) protocol (9). After extracting RNA, the quantity and quality of RNA were evaluated by UV spectrophotometry and agarose gel electrophoresis. In the spectrophotometric method, the concentration of RNA sample was measured using 260 nm wavelength absorption determination. The ratio of A260 / A280 was used to determine the purity of RNA. Samples with the appropriate OD (range 1.8 to 2) were selected for the next steps. In the electrophoresis method, 3 microliters of RNA dissolved in deionized water were used on 1% agarose gel. The presence of two 18S and 28S ribosomal bands indicated that RNA was purity. After extracting the appropriate quality RNA, it is necessary to convert it to cDNA to start RT-PCR. This was performed using the reversal transcriptase. For cDNA synthesize Pars Toos kit (Iran) was used. Thus, the cDNA generated was maintained at -20°C until the real time PCR was performed and expression of the NLRP1 gene was assessed (10).
Gene Expression
To investigate gene expression, Cyber-Green real time PCR was used. To design the primers, the site www.ncbi.nlm.nich.gov and the Primer Blast software were used to evaluate the capabilities of these primers based on their physical and chemical properties, the specificity of the primers, and the possibility of their compatibility with other organisms. The Oligo and Nucleotid Blast programs were then used for more accurate primer verification. Primers designed by Iran Takapozist Company were synthesized. The sequence of primers used for NLRP1: F: 5`- ACTCTCCCTCATTCTCCCCTAC -3`, R: 5`- GCTGTCTCAAAACACCCTTCTC -3`, and for beta actin: F: F:5`-GGCACCCAGCACAATGAAG-3`, R: 5`-CCGATCCACACGGAGTACTTG-3`. A suitable amount of deionized distilled water was added to the primers to obtain a 100 pmol/µL concentration. A certain volume was separated from the original solution and the residue was stored at -20°C. Before examining the gene expression, to evaluate the quality of the genes studied and also to determine the optimal value of the primer and the connection temperature of the primer, the molecular method of PCR with temperature gradient was used. Therefore, in each reaction, different concentrations of primer (5 and 10 picomoles) were used and the thermocycler system temperature gradient program was used to determine the optimal connection temperature. To perform PCR in a 0.2 mL sterile microtubule, the following ingredients were added (Table 1) and then the microtubes were placed in a thermocycler and the system was set up in accordance with Table 2 protocol. For gene expression, SYBR Premix Ex Taq II Kit (Takara, Japan) using the design and optimized primers according to the kit instructions was used (11).
| Concentration | (µl) | Materials |
| 1X | 2.5 | Buffer 10X PCR |
| 0.2 mM | 0.5 | dNTP 10nM |
| 1.5 mM | 0.75 | Mgcl2 50mM |
| 1.5 unit | 0.25 | Taq DNA Polymerase 5unit/μL |
| 1 μM | 0.5 | Forward primers 10μM |
| 1 μM | 0.5 | Revrese primers 10μM |
| --- | 15 | Distilled water |
| --- | 5 | cDNA |
| --- | 25 | Final volume |
Table 2. Time program of thermal cycler for NLRP1 and β-Actin
| No. of Cycles | Time | Tem(°C) | Stage |
| 1 | 5 min | 94 | Pre-denaturation |
| 35 |
1 min 55 Sec 1 min |
94 55-63 72 |
Denaturation Annealing Extension |
| 1 | 5 min | 72 | Final extension |
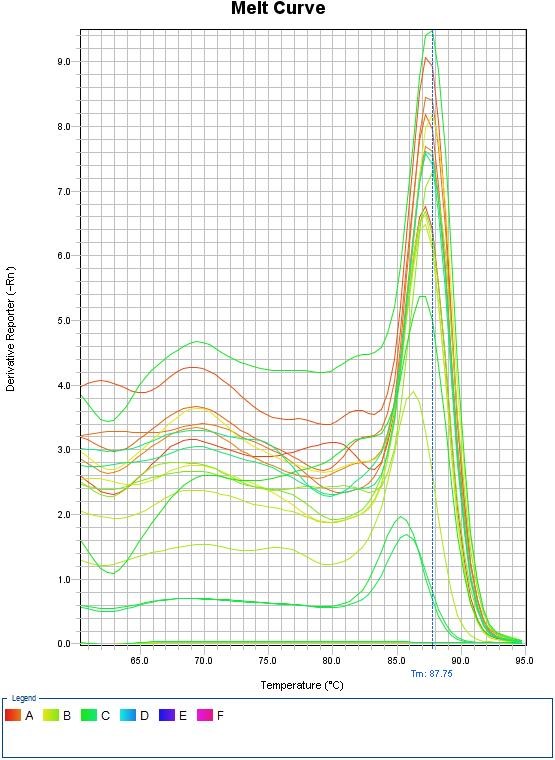
Melt Curve Analysis
By analyzing the melting curve, the existence of non-specific bonds and dimmer primers can be detected. In the real-time PCR method, in addition to controlling DNA synthesis during the reaction and drawing the multiplication diagram, the melting point of the product was determined at the end of the reaction and the melting curve was drawn. This was done after the PCR process was completed. All PCR products produced by a particular pair of primers have the same melting point, as Cybergreen is unable to distinguish between target DNA and other long-distance DNAs. Using the melting curve, the variety of products in the PCR process can be examined. The melting diagram was drawn for each sample by measuring the fluorescence changes at different temperatures. The sudden decrease in the intensity of the fluorescence that occurs with the opening of the DNA strand can be seen as a peak in this curve. Each peak represents Tm, a PCR product.
Data Analysis and Statistical Methods
The differences between patients with septicemia and healthy controls and female compared to male were calculated using Mann–Whitney U test, and the differences among the patients infected with various bacteria were examined using Kruskal–Wallis test using SPSS software version 18. P-value was considered significant at <0.05. Measurement of gene expression was measured using the conventional Livak method and with the evaluation of 2- DDC.
The results of blood culture were determined by observing the growth of bacteria. Escherichia coli: 7, Staphylococcus aureus: 10, Acinetobacter baumannii: 17 and Pseudomonas aeruginosa: 6 cases were responsible for septicemia in patients. The results of primer temperature optimization showed that the best temperature for connecting primer for NLRP1, beta-actin were 57°C and 58°C, respectively. The melting curve also confirmed the accuracy and specificity of the NLRP1 gene expression (Figure 1). Based on the obtained diagram, it was observed that these genes had only one peak in the temperature range of 85 to 90°C, which indicates that they are single-band and the specificity of the related reaction and non-contamination.
Figure 1. Melting curve of NLRP1 gene
Results of Inflamazome NLRP1 Gene Expression in Septicemia Patients and Control Group
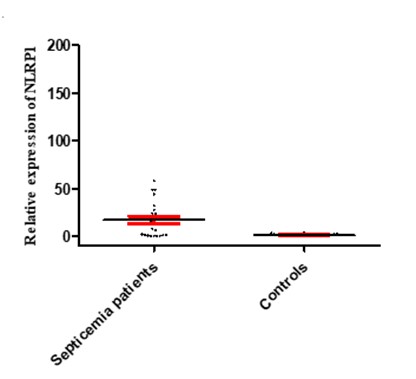
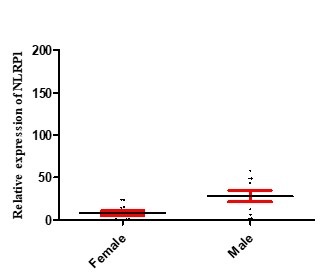
Inflamasome NLRP1 had a significant increase in expression in people with septicemia compared to healthy controls, as shown in Figure 2. As shown in Figure 3, the expression of the NLRP1 gene in men was higher than women. Figure 4 shows that there is no significant difference in the expression of NLRP1 gene expression in relation to the bacteria causing the infection isolated from septicemia samples such as Staphylococcus aureus, Escherichia coli, Acinetobacter baumannii and Pseudomonas aeruginosa.
Figure 2. Expression levels of NLRP1 in septicemia patients and healthy controls. The figure illustrates that the mRNA levels of NLRP1 was significantly increased in septicemia patients compared to healthy ones.
Figure 3. NLRP1 expression levels in male and female patients with septicemia. The results show that NLRP1 at the mRNA level significantly increased in male patients (28.06 ±6.76) compared to women (2.82 ±2.86) in septicemia patients (p = 0.011).
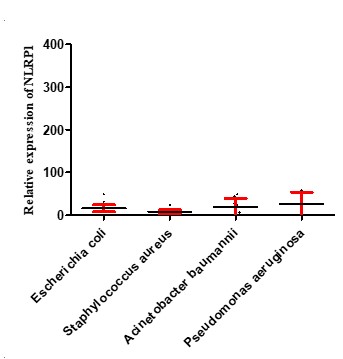
Figure 4. Expression levels of NLRP1 in septicemia patients infected with various bacteria. The figure illustrates that the mRNA levels of MDA5 and RIG-1 did not significantly differ among groups.
Septicemia is the third leading cause of death after lung infection and AIDS, and is the most common cause of death in the intensive care unit, given the growing trend and subsequent problems, including septic shock and in the last 20 years; the trend has been increasing (12). Septicemia caused by gram-negative bacteria is more deadly than other bacteria, but gram-positive bacteria, anaerobes, fungi, and even viruses can be among the etiologies of these bacteria (13, 14). At least part of the cause of death due to septicemia is due to a lack of specific clinical signs for early diagnosis. In addition, the absence of specific markers of infection in this disease has led to problems in differentiating the syndrome of systemic inflammatory response caused by infectious agents from non-infectious, which in turn leads to low consumption or in some cases high consumption of antimicrobial agents (15). The presence of clinical symptoms alone is not enough to diagnose septicemia and a prompt and appropriate diagnosis of toxic sepsis should be a daily challenge for the emergency department and the intensive care unit. Today, various treatments have improved the survival of patients with septicemia, so a prompt and correct diagnosis is essential. Septicemia has no specific symptoms, microbial culture requires time, and does not show a systemic inflammatory response. At the same time, it does not show a defect in the function of organs and may not yield positive results in some patients with septicemia for various reasons. Septicemia is a systemic reaction of the body to invasive microorganisms, including bacteria and fungi (16). About two-thirds of septicemia occurs in hospitalized patients, with a number of factors such as increasing population age, increasing duration of chronic disease, use of antibiotics and corticosteroids, and mechanical and intravascular devices to increase its incidence (1). Septicemia usually occurs in people with weakened immune systems (17). In the present study, similar to previous studies, the prevalence of toxic sepsis was determined, especially in hospitalized individuals, and this could be due to nosocomial infections, the use of catheters, and contaminated instruments. The highest levels of contamination in the present study were related to gram-negative bacteria, and 42.5% of the infections were caused by Acinetobacter baumannii. This bacterium is also one of the major causes of nosocomial infections, especially in the intensive care unit, and has a wide range of antibiotic resistance (27). Primary and innate immune responses play a key role in eradicating infections from tissues, including blood (18) and since NLRP1 and NLRP3, as intracellular receptors, play an important role in inducing primary immune responses; these molecules appear to play an important role in inducing primary immune responses against septicemia-causing bacteria (19). NLRP1 is one of the first known inflamasomes. In some bacterial infections, the path of pyroptosis is activated. ATP leakage from inflammatory macrophages and its entry into the cell through the canexin canal and ATP binding to the P2X7 receptor lead to NLRP1 activation. Decreased cellular ATP activates NLRP1. The two anti-apoptotic proteins Bcl-2 and Bcl-XL prevent the activation of NLRP1. NLRP1 ability to detect cellular energy levels may establish a link between metabolism and the immune system. Some studies have suggested that NLRP1 may be a sensor for hematopoietic stress and an important regulator of inflammation (6, 20). In the present study, NLRP1 also had a significant increase in expression in patients with septicemia compared with the control group. Investigations have shown that 3NLRP molecules play an important role in identifying and responding appropriately against intracellular infections (21). Other studies suggest that Inflamazom NLRP3 acts as part of the host defense against a large group of activated bacterial, fungal, and viral pathogens, producing IL-18 and IL-1 (22). Another study found that the innate immune system produces a set of cytoplasmic sensors that are activated against viral infections. Activation of these sensors leads to the production of interferon type 1, inflamasome activity, and the production of inflammatory IL-1 and IL-18 cytokines (23). Another study showed that inflamasomes play an important role in causing cirrhosis as well as liver cancer following liver inflammation (24). A study of patients with hepatitis C also found that NLRP3, as one of the inflamasomes, played an important role in activating IL-1 by identifying components of the hepatitis C virus (25).
NLRP1 appears to be an important receptor for bacteria during bacterial bloodstream infection, and further research, especially in reducing the expression of NLRP1 molecules, could play a key role in purifying the blood. Determine the cause of infection.
We thank from colleagues of the Microbiology Research Laboratory and the Research Department of the Islamic Azad University, Kerman Branch.
Authors declared no conflict of interests.
Received: 2020/02/28 | Accepted: 2020/06/16 | ePublished: 2020/05/12
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |