BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-987-en.html
2- Molecular Biology Research Center, Baqiyatallah University of MeIrandical of Sciences, Tehran, Iran
3- Department of Microbiology, Karaj Branch, Islamic Azad University, Karaj, Iran ,
.
Klebsiella pneumoniae (K. pneumoniae) is an opportunistic bacterium and an important pathogen is responsible for a wide range of nosocomial acquired from the hospital and is the cause of urinary tract infections, neonatal arthritis, meningitis, wound infections, nosocomial pneumonia, bacteremia, septicemia, and soft tissue infections (1). The microorganism is also a potentially pathogenic community-acquired pathogen (1). Most K. pneumoniae isolates are multidrug-resistant (2). Although fluoroquinolones such as ciprofloxacin are often used to treat K. pneumoniae infections, the resistance of this bacterium to this group of antibiotics is increasing (3, 4). The resistance to fluoroquinolones in gram-negative bacteria is often due to chromosomal mutations in the gyr and par genes (5, 6). This research aimed at investigating the pattern of fluoroquinolone resistance and its relation with a mutation in the parC gene among clinical isolates of K. pneumoniae.
In this descriptive-analytic study, 95 K. pneumoniae. Isolates were initially identified using standard laboratory methods including growth on MacConkey agar medium (Merck, Germany) at 37 ˚C, showing the purple appearance, Gram stain (Gram-negative coccobacillus), oxidase test (negative), sulfide indole motility (SIM), methyl red-Voges-Proskauer (MR-VP), citrate utilization, urease test, triple sugar iron (lactose fermentative or acid/acid, G+, H2S-) and for molecular diagnosis, Presence of 16S rRNA gene was verified using PCR (fig 1). The total genomic DNA was extracted from K. pneumoniae colonies grown on LB broth (Merck Co., Germany) by the boiling method. DNA quality and concentrations were determined by Nanodrop spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA) and agarose gel electrophoresis. Whole extracted DNAs were immediately stored at -70°C.
After biochemical and molecular diagnosis screened for resistance to fluoroquinolones and anther antibiotics by the Kirby-Bauer disc diffusion technique on Mueller-Hinton Agar (Merck Co., Germany) medium according to the Clinical and Laboratory Standards Institute 2018 (CLSI) strategies.
Table 1. Primers used inhis stu tdy
| Reference | Product size | Seq | Gene |
| (4) | 340 bp | F 5′-CAGCTCGGCATACTTCGAC-3′ R 5′-CCTGAACTACTCCATGTACGTGAT-3′ |
parC |
| (15) | 130 bp | F 5′-ATTTGAAGAGGTTGCAACGAT-3′ R 5′-TTCACTCTGAAGTTTTCTTGTGTTC-3′ |
16S rRNA |
Table 2. Thermocycler PCR protocol for the 16S rRNA gene
| Time | Temperature | Steps |
| 4 min | 94˚C | Initial Denaturation |
| 45 sec | 94˚C | Denaturation |
| 45 sec | 59˚C | Annealing |
| 45 sec | 72˚C | Extension |
| 10 min | 72˚C | Final Extension |
| Time | Temperature | Steps |
| 4 min | 94˚C | Initial Denaturation |
| 30 sec | 94˚C | Denaturation |
| 30 sec | 50˚C | Annealing |
| 1 min | 72˚C | Extension |
| 7min | 72˚C | Final Extension |
Determination of MIC of ciprofloxacin by microbial dilution method
The MIC of ciprofloxacin was measured by dilution microbial assay at concentrations ranging from 0.25 - 1024 µg / ml.
PCR assay for detection of parC genes
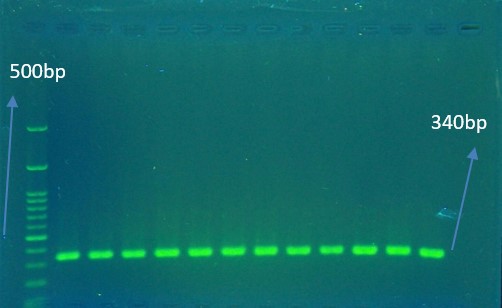
The mutation in the determining area for resistance to ciprofloxacin in the parC gene of resistant K. pneumoniae was performed by the reproduction of this gene using the PCR method and nucleotide sequencing analysis (Figure 2 ,3 ,4 5).
The desired genes and their primer sequences used in our study were demonstrated in Table 1. The total volume of reaction per each PCR test was 25 µL in the PCR tube. The reaction mixtures for detection of parC gene contained 1 μL DNA (10 pmol/L), 12.5 μL ready 2× PCR Master Mix (SinaClon BioScience Co., Iran), 1 μL of 10 pmol/L of each primer and 9.5 μL of sterile ultrapure water up to 25 µl volumes. Thermal cycling conditions for gene amplification were performed in Table 2 & 3. Finally, the effect of the mutation on the structure of IV topoisomerase enzyme and its possible role in resistance to Ciprofloxacin was investigated using online Blast, Insilico, and Clustalw2 software.
Sequencing analysis showed that 16 of 23 isolates resistant to ciprofloxacin had I80S mutations and an isolate also had an E84K mutation in the parC gene. According to the results, a mutation in parC gene is one of the most important mechanisms of resistance to fluoroquinolones in clinical isolates of K. pneumoniae. The mutation in the parC gene induces resistance to ciprofloxacin in K. pneumoniae by altering the tendency of ciprofloxacin to IV topoisomerase, and this can contribute to the increase of K. pneumoniae isolates resistant to common antibiotics and the increased incidence of nosocomial infections.
Table 4. The results of the antimicrobial susceptibility pattern of K. pneumoniae isolates
| Antibiotics | Susceptible | Intermediate | Resistance |
|---|---|---|---|
| Cefepime 30 μg | 67 (70.53%) | 1 (1.05%) | 27 (28.42%) |
| Tobramycin 10 μg | 72 (75.79%) | 8 (8.42%) | 15 (15.79) |
| Chloramphenicol 30 μg | 71 (74.74%) | 8 (8.42%) | 16 (15.79%) |
| Ciprofloxacin 5 μg | 61 (64.21%) | 11 (11.58%) | 23 (24.21%) |
| Nalidixic acid 30 μg | 56 (58.95%) | 13 (13.68%) | 26 (27.37%) |
| Trimethoprim-sulfamethoxazole 75 μg | 62(65.26%) | 0 | 33(34.74%) |
| Piperacillin 100 μg | 9 (9.48%) | 20 (21.05%) | 66 (69.47%) |
| Tetracycline 30 μg | 65(68.42%) | 6 (6.32%) | 24 (25.26%) |
| Imipenem 10 μg | 77 (81.05%) | 5 (5.26%) | 13(13.69%) |
| Cefecxim 5 μg | 57 (60%) | 3 (3.16%) | 35 (36.84%) |
| Meropenem 10 μg | 75 (78.95%) | 1 (1.05%) | 19 (20%) |
| Ampicillin 10 μg | 1 (1.05%) | 0 | 94 (98.95%) |
| Amoxicillin 10 μg | 1 (1.05%) | 0 | 94 (98.95%) |
DNA topoisomerase IV subunit A
Query 11 GCATGGCGGCG--AGATTTGGGATCGTCCGGCGCCCCCCAGTTTCCCTGACCATCCACCA 68
||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 701656 GCATGGCGGCGAAAGATTTGGGATCGTCCGGCGCCCCCCAGTTTCCCTGACCATCCACCA 701597
Query 69 GCGGATAGCGGTAAGAGAACGGCTGCGCCATCAGCACCATCGCTTCATAGCAGGCGATGT 128
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 701596 GCGGATAGCGGTAAGAGAACGGCTGCGCCATCAGCACCATCGCTTCATAGCAGGCGATGT 701537
Query 129 CGCCGTGCGGGTGATATTTACCCAACACGTCGCCGACAGTGCGGGCGGACTTTTTGAATT 188
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 701536 CGCCGTGCGGGTGATATTTACCCAACACGTCGCCGACAGTGCGGGCGGACTTTTTGAATT 701477
Query 189 TCGCGCTGGCGTTCAGCCCCAGCTCGGACATCGCATAGACGATGCGACGCTGGACCGGTT 248
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 701476 TCGCGCTGGCGTTCAGCCCCAGCTCGGACATCGCATAGACGATGCGACGCTGGACCGGTT 701417
Query 249 TTAAGCCATCGCCAATAAACGGTAATGCCCTGTCCATGATCACGTACATGGAGTAGTTCA 308
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 701416 TTAAGCCATCGCCAATAAACGGTAATGCCCTGTCCATGATCACGTACATGGAGTAGTTCA 701357
Query 309 GG 310
||
Sbjct 701356 GG 701355
Figure 3. Examining the presence of mutations in a fragment of a replicated parC gene and comparing it with the standard sample available on the NCBI site shows the color of the mutation site in code 80.
Figure 4. Chart for electrogram, one of the examples of sequence determination
EMBOSS_001 1 ----------------------MYVIMDRALPFIGDGLKPVQRRIVYAMS 28
||||||||||||||||||||||||||||
EMBOSS_001 1 MSDMAERLALHEFTENAYLNYSMYVIMDRALPFIGDGLKPVQRRIVYAMS 50
EMBOSS_001 29 ELGLNASAKFKKSARTVGDVLGKYHPHGDIACYEAMVLMAQPFSYRYPLV 78
|||||||||||||||||||||||||||||.||||||||||||||||||||
EMBOSS_001 51 ELGLNASAKFKKSARTVGDVLGKYHPHGDSACYEAMVLMAQPFSYRYPLV 100
EMBOSS_001 79 DGQGNWGAPDDPKS---RRHANQ--------------------------- 98
||||||||||||||.|:...
EMBOSS_001 101 DGQGNWGAPDDPKSFAAMRYTESRLSKYSELLLSELGQGTADWVPNFDGT 150
EMBOSS_001 99 -------------------------------------------------- 98
EMBOSS_001 151 LQEPKMLPARLPNILLNGTTGIAVGMATDIPPHNLREVAQAAIALIDQPK 200
EMBOSS_001 99 -------------------------------------------------- 98
EMBOSS_001 201 TTLDQLLDIVQGPDYPTEAEIITSRAEIRKIYENGRGSVRMRAVWKKEDG 250
Figure 5. Shows a comparison of the amino acid sequence of a gene amplified with a standard sample and the location of the amino acid change that the catheter shows instead of isoleucine instead of serine.
Klebsiella is the cause of a wide range of diseases and one of the most important bacteria that we have witnessed in recent decades due to the indiscriminate and unscientific use of antibiotics (3). Fluoroquinolones are a group of broad-spectrum synthetic antibiotics that are widely used in medicine due to their high oral absorption. However, the widespread use of these antibiotics has led to an increase of resistance to them in recent years (17-19). Enzymes, DNA gyrase, and topoisomerase IV, which are essential for replication and transcription, are the targets of fluoroquinolones. The resistance of gram-negative bacteria to fluoroquinolones is often the result of chromosomal mutations and the displacement of certain amino acids in the Quinolone-Resistance Determining Region (QRDR) (20, 21). This study aimed to investigate the pattern of resistance to fluoroquinolones and its association with mutation in the parC gene among the clinical isolates of K. pneumoniae. The results of this study showed an increase in the rate of resistance of K. pneumoniae isolates to various antibiotics, which is compatible with studies conducted by Ahanjan et al, and Hashemi et al (22, 23). The results of this research also showed that 72.6% of the isolates were ESBLs + and 70.5% of the isolates being studied were MDR and 8.4% of the isolates were XDR. No drug-resistant isolate (PDR) was observed in this study; The results of the significant increase in ESBLs + and MDR isolates compared to Pourali Sheshblouki et al study in 2016 (24), but the results of Shivaee et al study are consistent (25). The results also showed that from 95 isolates of K. pneumoniae, 24.2% were resistant to ciprofloxacin; Which was almost consistent with the studies conducted by Norouzi et al in 2014 in Kerman and a study by Pourali Sheshblouki et al in 2016 (24,26). But the results were not consistent with Molana et al's study in 2010 and Mohammad Alipore et al's study in 2013 in Tabriz (4,27). This can be due to the different distribution of infections in different provinces, genetic diversity, differences in the location of isolates, the number of isolates, the type of samples collected, the type of discs used, or personnel errors could be the reasons for the differences in various studies. Various studies have suggested a variety of mutations, including mutations in codons 80 and 84 in the parC gene, which include S80I and E84K, and major causes of resistance to ciprofloxacin. The results of the present study showed that all 23 studied isolates in the parC gene had mutations at one or more points, and a comparison of the nucleotide sequence of the parC gene with the parC gene of the standard K. pneumoniae ATCC13883 showed that the mutation changed the amino acid sequence at 20 isolates from 23 being researched isolates; Of these, 16 mutations were observed in codon No. 80, which resulted in the conversion of serine (S) to isoleucine (I), and in a sample in codon No. 84 in which amino acid E (glutamic acid) was mixed with the amino acid. K (lysine) was displaced, 3 mutations caused a change in the framework and a wide change in the sequence of the enzyme topoisomerase IV, and in three samples the mutation did not change the amino acid sequence; Therefore, the most common mutation was codon No. 80, which was caused by isoleucine mutation instead of serine. However, the conversion of glutamic acid to lysine amino acids, alanine, and glycine in various studies, including the study of Minarini et al, in Brazil between 2002 and 2005 (27), Brisse et al, in the Netherlands in 2001 (29), Deguchi et al, 1996 in Japan. (3) was observed. In the study of Brisse et al, several mutations, including S80I and E84K, were observed in the isolates of K. pneumoniae (29). In a 2015 study by Piekarska et al, both mutations were detected in fluoroquinolone-resistant isolates (30). In the Minarini et al study, S80I mutations were identified in six of the 21 quinolone-resistant isolates obtained, but no E84K mutation was reported (28). In a 2003 study of Chen et al, no mutations in the parC gene were found among the 34 resistant strains of K.pneumoniae studied in Taiwan, but it was found that some resistant strains had mutations in gyrA (32). In the study of Mohammad Alipore et al, in 2015, on 10 isolates of Cibrosila pneumoniae resistant to ciprofloxacin, six isolates had mutations in the gyrA gene (27). In the study of Park et al in 2017, of the 42 K. pneumoniae-resistant Ciprofloxacin isolates, 36 isolates had at least one mutation in one of the gyrA, gyrB, and parC genes (31). In various studies such as the study of Brisse and Piekarska, E84K mutations were reported in some fluoroquinolone-resistant isolates (30, 29). In the Norouzi et al study, 6 out of 111 K. pneumoniae isolates had mutations in the parC gene that had only two common S80I mutations but no E84K mutations (26). The E84K mutation is one of the most common mutations in parC that has been reported in fluoroquinolone-resistant isolates. The occurrence of this mutation in a ciprofloxacin-resistant K. pneumoniae isolates in this study indicates its importance in drug resistance. Most of the results studied and also the results of this study show that point mutation in codons 80 and 84 of the parC gene is present in most samples resistant to ciprofloxacin, and in the meantime, point mutations in codon 80 are more important it has.
Drug resistance is still increasing in K. pneumoniae. Considering that ciprofloxacin is one of the effective antibiotics in the treatment of nosocomial infections caused by K. pneumoniae, the presence of 23 ciprofloxacin-resistant isolates from 95 isolates as well as resistance to high doses of ciprofloxacin antidepressants in this study increased the risk of silent resistance. Also, the occurrence of point mutations in the parC gene in all ciprofloxacin-resistant isolates indicates the importance of these mutations in the resistance to ciprofloxacin resistance in this bacterium.
This research article is taken from a Master’s Degree in Microbiology.
The authors of this article are grateful to the Honorable President of Molecular Laboratory and Expert of Microbiology Research Laboratory, Islamic Azad University, Karaj Branch.
Authors declared no conflict of interests.
Received: 2019/10/29 | Accepted: 2020/03/21 | ePublished: 2020/05/12
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |