BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-2538-en.html

 , Parastoo Saniee2
, Parastoo Saniee2 

 , Mohaddaseh Ramezani3
, Mohaddaseh Ramezani3 
 , Paria Ghadersoltani1
, Paria Ghadersoltani1 
 , Mohammadreza Najafi-disfani1
, Mohammadreza Najafi-disfani1 

2- Department of Microbiology and Microbial Biotechnology, Faculty of Life Sciences and Biotechnology, Shahid Beheshti University, Tehran, Iran ,
3- Microorganisms Bank, Iranian Biological Resource Centre (IBRC), Academic Center for Education, Culture and Research (ACECR), Tehran, Iran
The human body harbors a diverse array of microbiota, comprising a vast population of microorganisms that coexist in symbiosis with multicellular organisms (1). It is suggested that the number of microorganisms is 10 times more than the number of human cells in human body and the amount of genes encoded by these microorganisms exceeded the human genes, with an estimated 150-fold increase (2). This incredible diversity has led to a re-evaluation of the human body as a complex, meta-community composed of both human cells and microbial symbionts (3, 4). Microbiota is harbored in various parts of the human body, with majority residing in digestive system, exerting a profound impact on human health by engaging in various physiological processes (5).
In recent years, there has been a growing interest in the relationship between human microbiome and diseases. Studies have led to the discovery that the human microbiome plays a crucial role in maintaining homeostasis and an imbalance of this microbiome, known as dysbiosis, can have a negative impact on human health even in the early stages of life (6). Historically, the stomach was thought to be a sterile environment due to its acidic properties, leading to a long-held assumption that it was devoid of microbial activity (7). The landmark discovery by Marshall and Warren in 1983 of Helicobacter pylori (H. pylori) in the gastric mucosa of individuals with gastritis and peptic ulcer disease led to a change in our understanding of the human stomach microbial ecology (8) and prompted the hypothesis that various microbial communities can not only colonize but also adapt and persist within the stomach (9). Due to the relatively low biomass of gastric microbiota -estimated to be only between 10¹ and 10³ bacteria per gram of gastric content- there has been less focus on this area in previous research. This contrasts sharply with the jejunum and ileum, which contain about 10⁴ to 10⁷ bacteria per gram, and the colon, where the density reaches 10¹¹ to 10¹² bacteria per gram. As a result, the gastric microbiota has not been so extensively studied as its intestinal counterparts (10).
In the context of gastrointestinal histopathology, the stomach is susceptible to various diseases, which can be understood through the framework of the Correa cascade (11). This conceptual model outlines the progression of gastric disease from normal to neoplastic tissue, encompassing diverse phases of microbial alterations and interactions between microorganisms and their hosts (12). The initial stages of this cascade include chronic gastritis and atrophic gastritis, two prevalent gastrointestinal disorders characterized by inflammation and tissue damage. Chronic gastritis is marked by inflammation and erosion of the gastric mucosa, while atrophic gastritis is characterized by inflammation accompanied by the loss of oxyntic glands (13). Studies have indicated that in addition to H. pylori, the colonization of other bacteria can cause dysbiosis of the stomach microbiome, ultimately leading to the development and progression of gastric lesions (14, 15). For instance, the microbiome composition in patients with atrophic gastritis includes a diverse array of bacterial families, not only Helicobacteraceae but also Streptococcaceae, Fusobacteriaceae, and Prevotellaceae (16, 17).
To date, the majority of research on gastric bacteria has focused on metagenomics methods, with few studies using cultivation techniques (18, 19). A study implementing culture-based methods could reveal significant insights into the microbial landscape in gastric biopsies, highlighting the advantages of this approach in the light of specific research objectives. While metagenomics offer an overview of microbial diversity, it may overlook significant dynamics such as the presence of viable but non-culturable (VBNC) microorganisms that are essential for understanding disease mechanisms (20, 21). Isolating and characterizing distinct bacterial strains allow for the examination of their individual growth characteristics, including responses to varying oxygen levels and temperatures, which are critical for their survival in the gastric environment (22). Furthermore, this methodology affords the opportunity to correlate specific strains with clinical outcomes, thereby deepening the understanding of their roles in the Correa cascade. Culture-based methods enable the isolation of viable microorganisms, many of which remain undetectable through molecular techniques alone. Recent advancements in culturomics have demonstrated the ability to uncover previously unculturable taxa that play critical roles in host-microbial interactions (23). While metagenomics studies offer a broader view of microbial diversity and functional potential, they often struggle with identifying unclassified species and may lack the resolution to assess specific microbial interactions. Consequently, previous investigations have highlighted the complexity of gut microbial communities but have been limited in pinpointing live microorganisms that contribute to gastric health (24).
In Iran, despite the significant impact on gastric health and disease, the critical gastric microbiota has been overlooked. This study, which is part of a comprehensive investigation into various gastric disorders, was set to elucidate the composition of the microbial populations in the stomach microbiome of individuals with gastritis and atrophic gastritis through a hybrid approach combining culturing and molecular methods.
2.1 Study Population and Sampling
This study is part of an extensive investigation to examine 25 patients, as determined by the Cochran formula, across five distinct pathological stages (five patients per stage). In 2022, gastric biopsy samples were collected from the antral region of the stomach from the patients referred for endoscopic examination at the Digestive Disease Research Institute in Tehran, Iran. For this study, 10 patients were selected: 5 diagnosed with gastritis and 5 with gastric atrophy. Both endoscopic observation and histopathological examination were conducted for accurate diagnosis of gastric disease. Due to the invasive nature of biopsy collection and the associated risks, it is not ethically feasible to obtain biopsies from individuals without any known gastric complaints or risk factors. Therefore, a healthy control group was not included in this study.
Ethical approval was granted by the National Institute of Medical Research Development (NIMAD) (Approval number: IR.NIMAD.REC.1398.421) and informed consent was obtained from all participating patients. Additionally, information regarding age, sex, history of H. pylori infection, medications, treatments, and other relevant factors, was gathered through standardized questionnaires. The exclusion criteria were known gastric cancer cases, treatment with antibiotics, proton pump inhibitors (PPIs), histamine H2-blockers (H2B), non-steroidal anti-inflammatory drugs (NSAIDs), or anticoagulant medications within the past four weeks, or being pregnant.
Efforts were made to collect and standardize the history of antibiotic use among individuals, although accurate tracking of antibiotic consumption remains a challenge due to the widespread self-medication practices. The biopsy samples were collected using disposable forceps to avoid contamination from throat and mouth bacteria. A standardized process was applied to control infections and contamination throughout all stages of endoscopy and sampling (25, 26). Biopsy samples for culturomics were placed in sterile 2 ml vials containing transport medium of 0.16% agar + normal saline, while samples for metagenomics placed into sterile 2 ml vials with RNA later solution (27). To minimize the environmental contamination, all containers were sterilized before use. Samples were placed in additional sterile Falcon tubes and transported on ice to the Microbiology Laboratory at the Iranian Biological Resource Center, where they were stored at -80°C for further analysis.
2.2 Bacterial Growth and Identification
The gastric biopsies were defrosted, homogenized, and cultured in sterile BHI broth to enhance enrichment. The incubation period lasted between 48 to 72 hr under both aerobic and anaerobic conditions. Subsequently, the samples were cultured in three distinct media: BHI agar with 5% sheep-defibrinated blood, Mueller-Hinton agar, and de Man-Rogosa-Sharpe (MRS) agar, which served as enriched non-selective, non-selective, and selective media for lactobacilli cultivation, respectively, followed by incubation at two different oxygen levels (aerobic/anaerobic) and two sets of temperature (37°C/28°C). The protocols were optimized according to previous study (28). For aerobic culturing, laminar flow hoods were utilized, while gloveboxes were employed for anaerobic culturing. Isolated colonies were purified through successive sub-culturing on non-selective media, followed by examination of their morphological characteristics, including size, hemolysis, shape, and margin. Unique morphotypes were isolated and cultured on non-selective BHI agar with blood for further purification. After this initial culturing, the bacteria were stored at -20°C for a short period before DNA extraction. Subsequently, the samples were transferred -80°C for long-term preservation.
2.3 DNA Extraction and Polymerase Chain Reaction (PCR)
To identify the bacterial species, genomic DNA was extracted and purified using the Sinaclon DNA extraction kit (DNPTM), following the manufacturer's guidelines. The extracted DNA was then analyzed using spectrophotometry with NanoDrop spectrophotometer to evaluate its quality and quantity. Subsequently, the 16S rRNA gene was sequenced using universal primers 27F (5'-AGAGTTGATCCTGGCTCAG-3') and 1492R (5'-GGTTACCTTGTTACGACTT-3') (29). One-direction sequencing was employed to provide genus-level identification consistent with metagenomics analysis.
The PCR reaction program consisted of an initial denaturation step at 95°C for 3 min, followed by 25 cycles of denaturation at 95°C for 45 sec, annealing at 55°C for 45 sec, extension at 72°C for 1:30 min, and a final extension step at 72°C for 10 min. The PCR product was then analyzed using agarose gel electrophoresis and sent to FAZA Pajooh Company (Iran), for Sanger sequencing. The sequencing was performed in a single direction using primer 27F. The obtained sequences were analyzed using ChromasPro software, the BLAST tool (http://www.ncbi.nlm.nih.gov), and the EZBiocloud database (https://www.ezbiocloud.net).
2.4 Statistical Analysis
Chi-Square tests were conducted using Excel version 2021 to evaluate the association between bacterial genera and two conditions: gastritis and atrophy. For each bacterial genus, observed counts were compared to expected counts to calculate Chi-Square contributions. This statistic, along with the degrees of freedom, was utilized to calculate P-value with significant level set at 0.05.
The biopsies from 10 patients with gastritis and gastric atrophy with mean age of 53±13.5 years (ranged 17-71 years) comprised 6 females and 4 males. A total of 28 bacterial strains were identified with 13 and 15 strains isolated from patients with gastritis and gastric atrophy, respectively. The Gram staining revealed that the majority (86%) of the isolated bacteria exhibited Gram-positive characteristics and demonstrated varying levels of growth on alternative media. While all of them were capable of growth on Blood agar medium, only 18 isolates were able to grow on Mueller-Hinton agar and 14 isolates on MRS medium.
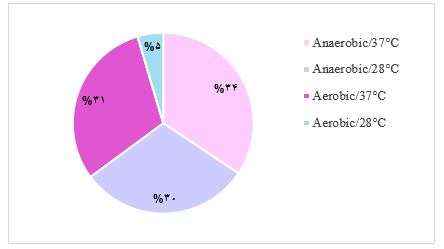
Furthermore, 64% of the isolated bacteria were able to grow in anaerobic conditions, whereas only 36% were capable of growth in aerobic conditions. The results also showed that 65% of the isolates demonstrated enhanced growth rate at 37°C, while only 35% were optimal at 28°C. Notably, the combination of anaerobic conditions at 37°C appeared to be the most favorable and aerobic conditions at 28°C represent the least favorable conditions for enriching bacteria from gastric biopsies (Figure 1).
Figure 1. The Percentage of isolated bacteria in various temperature and oxygen conditions
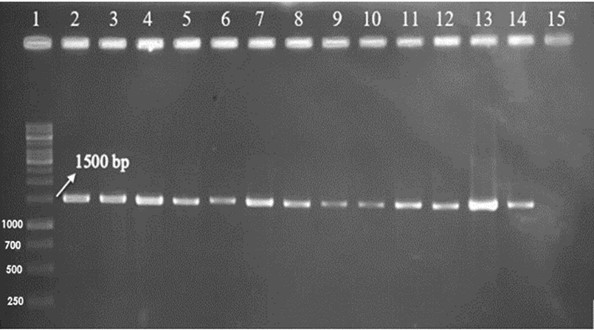
To further characterize the isolated bacteria, the prokaryotic 16S rRNA gene region was amplified using universal primers. The PCR products were visualized on agarose gel (Figure 2) and analyzed using ChromasPro software. A positive control, specifically Escherichia coli, was included in the experimental setup, although the positive control is not shown in Figure 2.
Figure 2. 16S rRNA amplification results: Well No. 1: 1kb ladder, Well No. 2-14: isolated bacteria, Well No. 15: negative control. The target band length was 1500 bp.
The extracted sequences were compared to GeneBank database to identify the bacteria at the genus level. Ten different bacterial genera were identified, with the highest percentage of identified bacteria belonging to the genus Bacillus (25%). The list of all identified bacteria is shown in Table 1.
Table 1. Classification of identified bacteria at the genus level
| Number | Percentage | Genus |
| 7 | 25% | Bacillus |
| 6 | 21.4% | Staphylococcus |
| 5 | 17.8% | Streptococcus |
| 3 | 10.7% | Lactobacillus |
| 2 | 7.1% | Stutzerimonas |
| 1 | 3.6% | Ligilactobacillu |
| 1 | 3.6% | Brevundimonas |
| 1 | 3.6% | Achromobacter |
| 1 | 3.6% | Lacticaseibacillus |
| 1 | 3.6% | Rothia |
| 28 | 100% | Total |
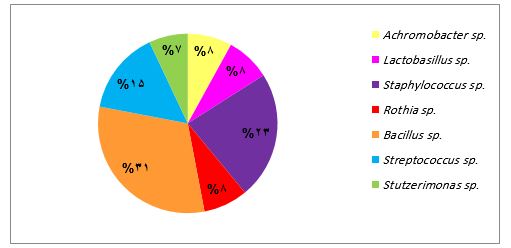
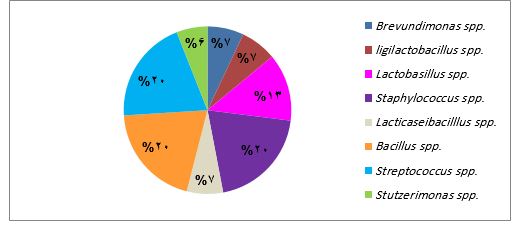
Furthermore, the analysis revealed a distinct bacterial diversity associated with gastritis and gastric atrophy. Achromobacter spp. and Rothia spp. were exclusively found in biopsies obtained from patients with gastritis (Figure 3), whereas Lacticaseibacillus spp., Ligilactobacillus spp. and Brevundimonas spp. were uniquely associated with gastric atrophy (Figure 4). In contrast, a shared bacterial community was observed among patients with both conditions.
While in gastric atrophy samples, the frequency of identified bacteria was evenly distributed among Bacillus spp., Streptococcus spp. and Staphylococcus spp. with a rate of 20%, in samples from patients with gastritis, Bacillus spp. was the most prevalent bacteria (31%), followed by Staphylococcus spp. (23%) and Streptococcus spp. (15%). This study further identified 10 distinct lactic acid bacterial strains, with 7 out of 10 being specifically linked to gastric atrophy patients (Table 2).
While the analysis indicates that certain genera are linked to different gastric conditions, the results of the Chi-Square test demonstrated no statistically significant association between the bacterial genera and the conditions of gastritis and atrophy (P=0.97 and 0.98, respectively).
Figure 3. Distribution of distinct bacterial genera isolated from biopsies obtained from patients with gastritis.
Figure 4. Distribution of distinct bacterial genera isolated from biopsies obtained from patients with gastric atrophy.
Table 2. The taxonomic classification of lactic acid bacterial strains isolated from 10 recruited patients.
| Order | Family | Genus | No. in patients with gastritis | No. in patients with atrophy | Total |
| Lactobacillales | Streptococcaceae | Streptococcus | 2 | 3 | 5 |
| Lactobacillales | Lactobacillaceae | Lactobacillus | 1 | 2 | 3 |
| Lactobacillales | Lactobacillaceae | Ligilactobacillus | 0 | 1 | 1 |
| Lactobacillales | Lactobacillaceae | Lacticaseibacillus | 0 | 1 | 1 |
| Total | 3 | 7 | 10 | ||
We indicated important insights into the complex microbial community of the stomach. The most identified bacteria were Bacillus, Streptococcus, Staphylococcus, and Lactobacillus detected in both gastritis and gastric atrophy biopsies. Lactic acid isolates were also found in association with patients suffering from gastric atrophy.
The human stomach microbiota has recently gained significant attention due to its potential influence on gut health and disease. Until the discovery of H. pylori in the early 1980s, it was believed that gastric acid, mucus layers, peristaltic motions, and the presence of antimicrobial factors inhibited the growth of microorganisms (16). However, the subsequent research revealed that in addition to H. pylori, other microorganisms were able to withstand the acidic environment of the stomach and colonize in this niche, causing diseases such as gastritis and gastric atrophy with significant contribution to dysbiosis (14, 30, 31). Elucidating the relationship between these microorganisms and gastric diseases can significantly advance our understanding of the mechanism of action and treatment strategies (16). The integration of conventional cultivation methods with molecular techniques, which is called culturomics, offers profound advantages. Culture-based methods not only enable the isolation and identification of viable microbial species but also capture their functional potential (21). By characterizing microbial communities in healthy and diseased gastric tissues, these methods facilitate insights into the host-bacterium interactions and the biological activities of bacterial products that may possess therapeutic potential (21, 32).
In the present study, the bacterial populations in 10 gastric biopsies obtained from patients diagnosed with gastritis and gastric atrophy were isolated and identified. The majority (86%) of the isolated bacterial strains exhibited Gram-positive phenotype, which is characterized by the presence of a thick peptidoglycan cell wall providing structural strength and rendering them more resistant to environmental stressors such as temperature and pH fluctuation (33). Furthermore, findings indicate that the majority of isolated bacteria were able to grow in anaerobic conditions. This suggests that the isolated bacteria possess a degree of adaptability to varying oxygen availability conditions, aligning with previous research that gastric bacteria are predominantly anaerobic and capable of tolerating oxygen to some extent (34, 35). Notably, the number of bacteria cultured at 37°C surpassed those grown at 28°C, likely due to the human body natural temperature providing more favorable environment for gastric microbiota growth and activity (36). Moreover, the isolated bacteria demonstrated varying levels of growth on alternative media with BHI agar medium being the most favorable one, highlighting different metabolic profile and adaptability to different environmental conditions (37).
In this study, 28 bacteria were identified as the most prevalent species belonging to the Bacillus genus, followed by Streptococcus, Staphylococcus, Lactobacillus, and Stutzerimonas. Staphylococcus aureus is known to exacerbate inflammation and mucosal damage during gastritis, potentially leading to gastric atrophy. In contrast, Staphylococcus epidermidis may exert a protective effect by outcompeting pathogenic bacteria and contributing to a balanced gastric microbiota (38). Similarly, Lactobacillus salivarius has been shown to mitigate gastric inflammation by regulating local cytokine secretion, suggesting a protective role against gastritis. Moreover, various strains such as Lactobacillus rhamnosus and Lactobacillus gasseri promote ulcer healing (17). Furthermore, certain Streptococcus species, including Streptococcus anginosus, are prevalent in gastric cancer patients compared to those with chronic gastritis, indicating a potential role in the progression from benign inflammation to malignancy. Conversely, Bacillus species, as normal components of gastric flora, contribute to maintaining microbial balance; however, their role may shift during dysbiosis states, potentially affecting gastric mucosal health (39).
Notably, in this study, a few bacteria were exclusively detected in biopsy samples from patients with gastric atrophy, specifically Ligilactobacillus, Brevundimonas, and Lacticaseibacillus. In contrast, Achromobacter and Rothia were exclusively found in biopsy samples from patients with gastritis. Potential mechanisms that may contribute to this specificity include modulating inflammatory responses, enhancing gastric mucosal integrity, and maintaining a balanced gut ecosystem. For example, Ligilactobacillus salivarius has shown anti-inflammatory effects by reducing pro-inflammatory cytokines and raising protective factors, indicating its potential therapeutic role in managing gastric atrophy (40). Additionally, a significant association has been found between Rothia mucilaginosa and chronic gastritis, indicating its potential role as a gastric pathogen alongside H. pylori. The presence of R. mucilaginosa was notably higher in gastric juice from chronic gastritis patients compared to controls, suggesting it may contribute to gastric inflammation (41). Further research is needed to explore the roles of various bacterial species in gastric conditions.
These findings reveal a diverse bacterial community in gastric microbiota aligning with several previous studies that have employed a combination of culture-based and molecular approaches to shed light on the gastric microbiota composition and potential links to stomach diseases (15, 17). It also reveals the probiotic properties of microbiota, which not only inhibit the colonization of pathogenic bacteria but also enhance anti-tumor immune responses (42, 43). A study conducted by Delgado et al., analyzed biopsy samples from 12 healthy individuals using both culture-dependent and pyrosequencing methods, revealing significant inter-individual variations in the microbiota community (44). Another study by Engstrand et al. (2013) showed that a healthy stomach is typically characterized by the dominance of specific bacterial genera, including Veillonella, Prevotella, Streptococcus, Clostridium, and Lactobacillus. The study also examined the gastric microbiota in individuals with atrophic gastritis, revealing a significant reduction in H. pylori abundance and an increase in other bacterial species. Furthermore, the presence of non-H. pylori bacteria in the stomach was associated with decreased gastric acid secretion and elevated risk of gastric cancer (45). Another study conducted by Ianiro et al., employed culture-independent analytical methods to investigate the gastric microbiome. The results indicated that human stomach is home to a diverse array of microbial communities, in addition to H. pylori, which exhibit both antibacterial and probiotic properties. These microbial communities may have therapeutic potential for the management of gastric diseases, offering new avenues for the treatment of gastric disorders (46). Moreover, some multi-omics analysis approaches, which included techniques such as genomics, proteomics, and metabolomics, demonstrated that composition of the gut microbiome is associated with the development and progression of gastric disorders, including chronic gastric atrophy, metaplasia, and gastric cancer (47, 48). These findings highlight the complexity of gastric microbiota and its significant implications for understanding gastric diseases, paving the way for future research aimed at harnessing the therapeutic potential of beneficial microbial communities (49).
Furthermore, 10 specific types of bacteria, known as lactic acid bacteria were isolated, which are commonly found in people with gastric atrophy. While these bacteria exhibit beneficial properties that suggest they could be potential candidates for probiotic use, it should be noted that extensive safety and supplementary tests are required to confirm their suitability for this purpose. The findings also indicate a strong association between these microorganisms and the development of gastric atrophy, warranting further investigation to explore their probiotic potential. This is consistent with previous research by Li et al., which found while Lactobacillus is often regarded as beneficial due to its probiotic properties, its overgrowth in gastric conditions is harmful. The reduced gastric acid secretion, typical in atrophic conditions, creates an alkaline environment conducive to Lactobacillus overgrowth. This increased abundance may disrupt the gastric microbiome balance, potentially facilitating dysbiosis that can promote inflammation and enhance carcinogenic processes. Additionally, Lactobacillus can modulate immune responses, sometimes leading to an inflammatory microenvironment, while its metabolic byproducts, such as lactic acid and nitrite from nitrate reduction, may further support tumor cell proliferation. While it is possible that Lactobacillus is simply a marker for bacterial overgrowth and not directly responsible for the development of gastric cancer, further research is needed to determine its exact role (50).
Although the statistical analysis showed no significant association between bacterial genera and the gastric conditions of gastritis and atrophy, it is important to note that some bacterial genera were identified as specific to different gastric conditions. This lack of significance might be due to factors like a small sample size or the absence of a healthy control group. Despite this limitation, the results were consistent with previous studies, suggesting that these bacterial genera may be relevant to these gastric conditions.
Integrating culturomics data with metagenomics studies can significantly enhance our understanding of the gastric microbiome. In this study, culturomics identified specific bacterial taxa linked to gastritis and gastric atrophy. Meanwhile, metagenomics can reveal shifts in microbial communities associated with various diseases, such as increased presence of non-H. pylori bacteria correlating with higher gastric cancer risks. However, metagenomics alone does not specify which live bacterial strains are involved in these disease processes without validation provided by culturomics. By comparing these findings, insights can be gained into how specific culturable populations contribute to the broader metagenomics landscape, elucidating their roles in health and disease. A key limitation of this study is the absence of a healthy control group, as biopsies were collected solely from patients with suspected gastric disorders. Future research should include healthy controls to differentiate between microbiota associated with a healthy gut and those linked to gastric disorders. Additionally, larger sample size and appropriate controls are necessary to better understand the relationships between bacterial genera and various gastric conditions.
This study fills the knowledge gap by exploring the stomach microbiome, highlighting the importance of non-H. pylori microorganisms. Establishing a solid foundation through culture-based analyses paves the way for future studies that could incorporate metagenomics techniques to expand upon these findings and investigate the complex interactions within microbial communities associated with gastric disease. It may lead to significant advancements in our understanding of the microbiota role in health and disease, ultimately informing the development of innovative therapeutic strategies and novel probiotics, as well as biomarkers for diagnosis and monitoring.
We express our gratitude to all the patients who participated in this project. Additionally, we appreciate the staff at Shariati Hospital for their assistance in sample collection.
Ethical Considerations
All patients signed an informed consent, and the study was approved by the National Institute of Medical Research Development (NIMAD) (Approval number: IR.NIMAD.REC.1398.421).
Authors’ Contributions
M.K. contributed to the methodology, investigation and writing the original draft. P.S. contributed to conceptualization, validation, project administration, writing the original draft, review & editing. M.R. contributed to conceptualization, validation and project administration. P. Gh contributed to the methodology, review and edition of the manuscript. M.N.D contributed to the methodology. All authors have read and approved the final version of the manuscript. The final manuscript was read and approved by all of the authors.
This research was conducted without external funding. The authors acknowledge that no specific grant or financial support from any organization was received for the completion of this study.
Conflicts of Interest
This study was supported by the National Institute of Medical Research Development (NIMAD) Grant No. 984874.
The authors were not utilized AI Tools.
Received: 2024/12/17 | Accepted: 2025/03/1 | ePublished: 2025/03/30
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |