BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-2212-en.html
2- Department of Biological Science and Technology, Najafabad Branch, Islamic Azad University, Najafabad, Iran
3- Department of Biology, Faculty of Basic Sciences, Shahrekord Branch, Islamic Azad University, Shahrekord, Iran ,
Gastric cancer (GC) is a significant global health burden, accounting for a substantial number of cancer-related deaths each year (1). Among the various risk factors identified, Helicobacter pylori (H. pylori) has garnered attention due to its association with the GC development and progression (2). The H. pylori infection is a widespread chronic bacterial infection that affects over the half of the world's population (3). Although most infected individuals remain asymptomatic, some develop progressive gastritis, peptic ulcers, and eventually gastric cancer (4). The exact mechanisms by which H. pylori contributes to the GC are not fully understood (5). However, it is widely recognized that H. pylori can induce a persistent inflammatory response, disrupt cellular homeostasis, and promote the genetic and epigenetic alterations within the gastric epithelium (6).
MicroRNAs (miRNAs) are a class of small, non-coding RNA molecules that play crucial roles in the post-transcriptional gene regulation (7). They are approximately 22 nucleotides in length and are highly conserved across the species. MiRNAs are involved in a wide range of biological processes, including development, cell differentiation, proliferation, and apoptosis (8). They have also been implicated in various diseases including cancer, cardiovascular disorders, and neurological conditions. MiRNAs can function by binding to the 3' untranslated region (UTR) of the target messenger RNAs (mRNAs), leading to the mRNA degradation or translational repression (9). This interaction between miRNAs and their target mRNAs is guided by the sequence complementarity, with partial complementarity resulting in translational repression and near-perfect complementarity leading to the mRNA degradation (10).
MiRNAs play a pivotal role in gene expression regulation and modulate immune responses during infections. Numerous studies have shown that H. pylori infection induces changes in miRNA expression profiles within gastric epithelial cells and immune cells. Dysregulation of miRNAs during H. pylori infection can result in abnormal expression of the target genes associated with immune responses, inflammation, and cell proliferation (11-13). For example, H. pylori-induced upregulation of miR-155 has been associated with increased pro-inflammatory cytokine production (14), while downregulation of miR-146a has been implicated in chronic inflammation and gastric carcinogenesis (15, 16).
Understanding the intricate relationship between miRNAs, H. pylori infection, and gastric cancer holds great promise for improving the diagnosis, prognosis, and treatment strategies (17). To this aim we attempted to identify the differentially expressed miRNAs associated with H.pylori infection via in silico methods and further detect its connection to gastric cancer via quantitative PCR method.
Sample Collection and RNA Extraction
A total of 80 blood samples were obtained from Al-Zahra Hospital in Isfahan, Iran. Among these samples, 40 were individuals with gastric cancer of which 29 samples were tested positive and 11 were tested negative for H. pylori infection. Forty other samples from healthy controls comprised of 25 H. pylori positive and 15 H. pylori negative samples. The samples infected with H. pylori were confirmed using the stool PCR test, and the cancer status of the samples was confirmed by a pathologist. In order to investigate the effect of gene expression on this cancer, two healthy and disease samples were selected with no significant difference in age and gender.
Demographic data regarding the collected samples have been demonstrated in Table 1. The collection of blood samples was conducted in accordance with the guidelines provided by the Ethics Committee of Islamic Azad University, Shahrekord branch, with approval number IR.IAU.SHK.REC.1401.083. These guidelines adhered to the principles outlined in the 64th World Medical Association General Assembly of Helsinki declaration, which was amended in October 2013. Furthermore, informed consent was obtained from all individuals included in the study.
The extraction of total RNA was carried out using Trizol reagent from the blood samples (Promega Co., based in Madison, WI, USA), then, the cDNA synthesis was performed using miScriptII® RT Kit (QIAGEN, Germany). QRT-PCR amplification was conducted by SYBR® Premix Ex Tag™ II (Takara, Dalian, China) using Qiagen Rotor-Gene real-time PCR instrument. The cycling parameters were set according to the instructions provided by the Takara kit including: initial denaturation: 95°C for 30 sec, amplification (40 cycles) of denaturation: 95°C for 5-10 sec, and annealing and extension: 60°C for 20-30 sec. Data were analyzed based on the 2−ΔΔCt method and the statistical analysis was performed using Prism software. The U6 snRNA was used as the control housekeeping gene. The miRNAs primer sets (Catalog #BON209002, Bonbiotech, BONmiR, Iran) were used for the qRT-PCR.
In-silico DEG Analysis
DEG analysis was performed on the expression data (GSE19769) from GEO database (18) using limma package (19) in R-based software (version 4.2.1) to identify the miRNAs with significant differential expression between the H. pylori positive and control samples. The differentially expressed miRNAs were selected based on the predefined criteria including fold change, p-value and adjusted p-value. First, the raw miRNA expression data was preprocessed. Then, statistical analysis was conducted to identify differentially expressed miRNAs between the H. Pylori positive and control groups.
For this purpose, the limma package was applied. This package considers the distribution of miRNAs and applies the statistical linear model to assess the significance of differences in the expression levels. Adjustments for multiple testing, such as the Benjamini-Hochberg procedure, controls the false discovery rate (FDR). Differentially expressed miRNAs were then selected based on the combination of fold change and statistical significance. The thresholds for both parameters were fold change of ≥1 and p-value of ≤0.05, to identify the miRNAs with substantial expression differences.
To validate the results of DEG analysis, quantitative real-time PCR (qRT-PCR) was performed. This technique confirmed the differential expression patterns of the selected miRNAs and further validated their potential functional relevance. Finally, the top10 most expressed genes of the DEG analysis are reported in Table 2. The most upregulated and downregulated miRNAs along with their fold changes and statistical significance are presented in Table 3.
Table 1. Demographic data of the samples.
| Total | Cancer | Normal | P-value | 95% CI |
| H. pylori H. pylori + H. pylori - |
40 | 40 | ||
| 29 11 |
25 15 |
0.0064 |
57.17 to 83.89 16.11 to 42.83 |
|
| Age <50 >50 |
20 20 |
18 22 |
>0.9999 |
35.20 to 64.80 35.20 to 64.80 |
| Sex Male Female |
28 12 |
26 14 |
0.0166 |
54.57 to 81.93 18.07 to 45.43 |
| Stage I-II III IV |
10 21 9 |
- |
<0.0001 |
- |
Table 2. Differentially expressed miRNAs.
| Differentially expressed miRNAs | P-value | Adj P-value | Logfc |
| hsa-mir-196b-5p | 0.0361599 | 4.75E-03 | 4.13 |
| hsa-miR-302a* | 0.0148171 | 1.28E-03 | 3.98 |
| hsa-miR-523 | 0.0185678 | 1.67E-03 | 3.69 |
| hsa-miR-596 | 0.0186046 | 1.73E-03 | 3.35 |
| hsa-miR-571 | 0.0106511 | 8.05E-04 | 3.22 |
| hsa-miR-548b | 0.0201644 | 1.99E-03 | 2.95 |
| hsa-miR-384 | 0.0217376 | 2.27E-03 | 2.4 |
| hsa-miR-519a | 0.0200165 | 1.94E-03 | 2.39 |
| hsa-miR-223 | 0.0001424 | 1.02E-06 | 2.05 |
| hsa-miR-146b | 0.0003573 | 3.86E-06 | 1.93 |
| hsa-mir-153-3p | 0.0026733 | 1.37E-04 | -5.81 |
| hsa-miR-411 | 0.0005893 | 1.36E-05 | -5.96 |
| hsa-miR-204 | 0.0186046 | 1.74E-03 | -3.69 |
| hsa-miR-96 | 0.0309951 | 3.74E-03 | -3.34 |
| hsa-miR-383 | 0.0461288 | 6.75E-03 | -3.07 |
| hsa-miR-299-5p | 0.1342025 | 3.26E-02 | -2.32 |
| kshv-miR-K12-10a | 0.0986511 | 2.11E-02 | -2.53 |
| kshv-miR-K12-10b | 0.1235873 | 2.89E-02 | -2.38 |
| hsa-miR-373 | 0.1732536 | 4.74E-02 | -2.13 |
| hsa-miR-369-5p | 0.2835105 | 9.33E-02 | -1.77 |
Table 3. Most upregulated and most downregulated miRNAs.
| Differentially expressed miRNAs | P-value | Adj P-value | Logfc | Average expression |
| hsa-miR-196b-5p | 4.75e-03 | 0.0361599 | 4.13 | H. pylori positive :4.648 |
| H. pylori negative:0.7228 | ||||
| hsa-miR-153-3p | 1.37e-04 | 0.0026733 | -5.81 | H. pylori positive :0.22 |
| H. pylori negative:3.045 |
The DEG analysis identified the miRNAs that exhibited significant differential expression between the H. pylori positive and normal samples. Based on the statistical analysis, a subset of miRNAs showed significant differential expression. Among them, 239 miRNAs were found to be significantly upregulated, while 318 miRNAs were significantly downregulated in the H. pylori positive samples compared to the control samples. The differential expression of these miRNAs was determined based on the predefined criteria, including a fold change threshold of 1 and a statistical significance level of P=0.05. Figure S1 shows the volcano plot demonstrating the differentially expressed miRNAs.
Real-Time PCR Validation
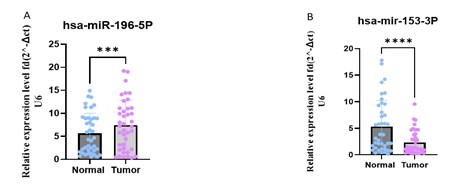
The most upregulated and the most downregulated miRNAs identified using real-time PCR were hsa-miR-196b-5p and hsa-miR-153-3p, respectively that were selected for the validation based on their LogFC and statistical significance in the DEG analysis. As shown in Figure 1, the real-time PCR results confirmed the significant upregulation of hsa-miR-196b-5p (P=0.0007) and significant downregulation of hsa-miR-153-3p in the tumor samples compared to the control samples (P<0.0001).
The hsa-miR-196b-5p exhibited 17.5 fold higher expression levels, indicating their increased abundance in the H. pylori condition. On the other hand, hsa-mir-153-3p showed 11.62 fold lower expression levels in the H. pylori positive samples relative to the control samples (Figure 2).
Statistical analysis was performed on the real-time PCR data to evaluate the significant level of the differential expression. The results revealed a significant difference (P<0.05) in the expression levels of the validated miRNAs between the H. pylori positive and control samples. No significant association was found between the age, sex, and disease stage, and the relative expression of hsa-miR-196b-5p or hsa-miR-153-3p.
Also, receiver operating characteristic (ROC) analysis was conducted and the ROC curve was illustrated in Figure 3.
Figure 1. Relative expression levels of hsa-miR-196b-5p and hsa-miR-153-3p in tumor vs. normal samples. (Designed by Authors, 2024)
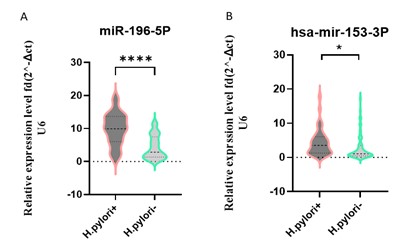
Figure 2. Relative expression levels of hsa-miR-196b-5p and hsa-miR-153-3p in H. pylori + vs. H. pylori – samples. (Designed by Authors, 2024)
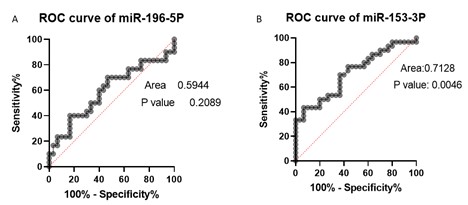
Figure 3. ROC curves of hsa-miR-196b-5p and hsa-miR-153-3p. (Designed by Authors, 2024)
The integration of differential expression analysis of GEO microarray data with real-time PCR validation has provided valuable insights into the regulatory landscape of miRNAs in H. pylori infection. In this study, we successfully identified the most significantly up-regulated (hsa-miR-196b-5p) and down-regulated (hsa-miR-153-3p) miRNAs using in-silico analysis, then, validated the expression of these miRNAs using real-time PCR. We demonstrated the association of different expressions of the hsa-miR-196b-5p and hsa-miR-153-3p with gastric cancer and H. Pylori infection.
The findings highlight the importance of integrating computational analyses with experimental validation to enhance our understanding of gene regulation in H. pylori infection, which in turn can lead to the gastric cancer (20, 21).
Some studies indicated the association between the gene expression and methylation patterns in gastric cancer (22). Particularly, this research has underlined the involvement of H. pylori infection in this connection. The findings of previous studies have demonstrated the complex and dynamic relationship between these factors, contributing to a better comprehension of the molecular mechanisms underlying gastric cancer (23). Mojtahedi et al., reported the association of iceA and vacA allelic genes with H. pylori strains and gastric disorders (24). In another study, Reyes et al., showed that H. pylori infection induces chronic inflammation that affects the gastric epithelium, which can lead to DNA damage and the promotion of precancerous lesions (25). Yao et al., identified novel diagnostic or prognostic markers for the gastric cancer through miRNA profiling using high-throughput sequencing or microarray technology. By means of this approach, they were able to identify miRNAs that showed differential expression in gastric cancer compared to the normal samples (26).
Among the validated miRNAs, hsa-miR-196b-5p demonstrated significant upregulation, suggesting its potential role as a positive regulator in H. pylori infection. These findings are in line with previous studies that have implicated hsa-miR-196b-5p in similar biological processes or disease contexts. In this regard a study has indicated that loss of TFF1 promotes the aberrant overexpression of HOXA10 and miR-196b-5p by demethylation of the HOXA10 promoter, which implies the cross-talk between the epigenetics and miRNA expression in human gastric cancer.
The overexpression of both miR-196b and HOXA10 was associated with the advanced tumor stage, lymph node metastasis, and poor overall survival in gastric cancer patients (27). Another study using high-throughput sequencing and microarray technology demonstrated that the differential expression of the hsa-miR-196b-5p was associated with esophageal cancer (28).
Conversely, hsa-miR-153-3p exhibited a significant downregulation, indicating its potential role as a negative regulator in H. pylori infection. The downregulation of hsa-miR-153-3p suggests that it may suppress the specific target genes or pathways implicated in H. pylori infection. Gao et al., have shown the ability of FGD5-AS1 as a means of controlling the functions of human gastric cancer cells, potentially by influencing the epigenetic axis of hsa-miR-153-3p-3p/CITED2 downstream, thus linking noncoding RNA-mediated carcinogenesis to epigenetics (29). In another study, the role of two microRNAs; miR-7 and miR-153 was explored in the development and progression of gastric cancer induced by H. pylori CagA protein.
The researchers found that both miR-7 and miR-153 are downregulated in the gastric cancer tissues, suggesting their involvement in the disease. The experimental evidence showed that CagA protein contributes to the downregulation of these microRNAs, which in turn leads to the increased proliferation, invasion, and migration of the gastric cancer cells. The findings highlight the potential of miR-7 and miR-153 as the therapeutic targets and provide insights into the mechanisms underlying H. pylori-induced gastric carcinogenesis (30).
By integrating the computational analysis and experimental validation, our study has contributed to the growing body of knowledge regarding the non-coding RNA-mediated regulatory mechanisms underlying H. pylori infection (31). The identified miRNAs, hsa-miR-196b-5p and hsa-miR-153-3p, hold promise as potential biomarkers or therapeutic targets in H. pylori infection. The validation of these miRNAs strengthens their candidacy for further investigation and exploration in the future studies. It is noteworthy that the mentioned miRNAs are differentially expressed in H. pylori condition, therefore it is deductible from this study results that the mentioned biomarkers may play a crucial role in gastric carcinogenesis.
It is important to note that the present study has certain limitations. Firstly, our analysis was limited to the available GEO microarray data, and additional experiments utilizing other datasets or platforms could further validate and expand our findings. Secondly, while real-time PCR is a robust and widely used technique for the gene expression analysis, it is essential to acknowledge that it has its own limitations, including the potential for the primer bias and variability in the amplification efficiency. Future studies could consider employing alternative validation methods such as RNA sequencing (RNA-seq) or functional assays to complement and verify our findings.
In conclusion, the integration of DEG analysis of GEO microarray data with real-time PCR validation has unveiled crucial miRNAs and provided valuable insights into their potential roles in H. pylori infection. The findings contribute to the growing body of knowledge in the field of H. pylori infection and pave the way for the future research endeavors aimed at therapeutic interventions and personalized medicine. Further investigations and functional studies are required to fully comprehend the regulatory mechanisms and clinical implications of these miRNAs in H. pylori infection. The comprehensive understanding of the miRNA-mediated regulation in H. pylori infection will ultimately contribute to the development of novel therapeutic strategies and improve the patients’ outcomes.
The authors would like to express their gratitude to all colleagues who have contributed to the completion of this research project.
Ethical Considerations
The collection of blood samples was conducted in accordance with the guidelines provided by the Ethics Committee of Islamic Azad University, Shahrekord branch, with approval number IR.IAU.SHK.REC.1401.083. These guidelines adhered to the principles outlined in the 64th World Medical Association General Assembly of Helsinki declaration, which was amended in October 2013. Furthermore, informed consent was obtained from all individuals included in the study.
Authors’ Contributions
The authors wish to declare their equal contribution for this research paper. All authors, have significantly contributed to the conception, design, and execution of the study. Throughout the research process, they jointly collected, analyzed, and interpreted the data. Their collaborative effort was essential in drafting and revising the manuscript, ensuring a cohesive and comprehensive presentation of the findings.
This research was conducted without external funding. The authors acknowledge that no specific grant or financial support from any organization was received for the completion of this study. The independence afforded by the absence of external funding underscores the authors' commitment to impartiality and objectivity in the research process.
Conflicts of Interest
Received: 2024/04/3 | Accepted: 2024/08/1 | ePublished: 2024/08/18
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |