BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-2169-en.html
2- Department of Microbiology, Shahr-e-Qods Branch, Islamic Azad University, Tehran, Iran ,
3- Department of Food Hygiene and Public Health, Faculty of Veterinary Medicine, Shahrekord Branch, Islamic Azad University, Shahrekord, Iran
4- Cellular and Developmental Research Center, Faculty of Basic Sciences, Shahrekord Branch, Islamic Azad University, Shahrekord, Iran
Functional foods and drinks derived from fermentation account for a large portion of the human diet and are believed to provide health benefits beyond essential nutrition (1). Fermentation has been employed since the dawn of time to extend the shelf life of natural resources. Also, it has improved the nutritional and sensory qualities of the finished product (2). Lactic acid bacteria (LAB) play the most critical role in fermentation (3). Also, these are the most common bacterial families found in processing a wide range of dairy products, particularly yogurt (4). The chemicals produced by these bacteria include peptides, amino acids, aldehydes, alcohols, organic acids, esters, and fatty acids. These chemicals are derived from the metabolism carried out by the LAB of the food components. In addition, they influence the shelf life, fragrance, taste, and texture of fermented foods (5). As a result, LAB is frequently employed as starters in food manufacturing operations (6). Clinical research has proven the beneficial effects of probiotic bacteria on enteric pathogens, allergic disorders, cancers, particularly clone cancer, digestion, and immune system modulation (7). Also, fermented yogurt, grains, and other sub-cultures are helpful products for enhancing health in many cultures. At the same time, large portions of them have yet to be researched, and the health claims have yet to be adequately supported by solid scientific data (8-11). LAB's technological features can benefit their industry sectors (12, 13). Besides, the technical potential consists of critical qualities for bacterial survival and the synthesis of chemicals that impact product attributes (14, 15). The Chaharmahal and Bakhtiari Province yogurt is one of these products (16). It is a concentrated form of milk's nutritional components that play a vital role in human nutrition (17-19). Various LABs, particularly the Lactobacillus genus, have been discovered while preparing traditional Chaharmahal and Bakhtiari Province yogurt (20, 21). Therefore, one of the objectives of the current study is to isolate LAB bacteria with probiotic potential for industrial applications. The present study examined LAB's probiotic and technical capabilities isolated from local yogurt in Chaharmahal and Bakhtiari Province, Iran.
2.1. Isolation and Identification
A) Sample Collection
Six samples of local yogurt were produced at a low temperature under sanitary conditions to avoid secondary infection or changes in the original microbiota. Sample collecting was done in 2022. In this case, 5 g of each sample was combined with 45 mL of sodium citrate (Merck, Germany). Then, it was homogenized with a stomacher to extract LAB strains (Interscience, France). On Man Rogosa and Sharpe agar (MRS agar), serial dilutions up to 10–7 were produced, and culture was performed (Conda, Spain).
B) Biochemical and Microbial Test Sample Identification
The samples were incubated at 37°C for 48 hours in an anaerobic environment (Gas-pack system, AmitisGen, Iran). Creamy to white, spherical, and somewhat slimy colonies were isolated. Gram-staining, catalase screening, and biochemical tests (Glucose fermentation, NH3 from arginine, Production of CO2 from Glucose, Dextran from sucrose, Acetate from glucose, Indole and oxidase test) and Growth at different temperatures 15˚C and 45˚C was done (22).
C) Molecular Identification
DNA was extracted from each representative sample. DNA extraction was performed using the Sinaclon kit (Sinaclon, Iran). On the consensus sequence of the 16S rDNA gene, Polymerase chain reaction was carried, (F: CTTGTACACACCGCCCGTCA; R: CTCAAA ACTAAACAAAGTTTC), (F: TGATGCATAGCCGAGTTGAG ; R: AACCGCCTGCACTCTCTTTA) and (F: TTCATGT AGGCGAGTTGCAG; R: ATTCGAAGCTACGCGAAGAA) and universal primers, 27FYM, and 1492R (23, 24). Subsequently, the amplified region was sent to the Sinacolon Industry in Iran for Sanger sequencing. Finally, the BLAST tool has been employed to compare data values with GenBank information (22).
2.2. Analysis of LAB probiotic properties
2.2.1. Bile Salt and pH Tolerance
Each isolate's capacity has been studied to develop on low pH and bile salt medium (Oxgall; Himedia, Mumbai, India). All isolates have been inoculated in MRS broth with 0.3% and 0.45% bile salt, respectively. The samples were incubated at 37°C for 24 hours. Then, the strains' development was monitored at 600 nm (25, 26).
2.2.2. Analysis of Resistance to Simulated Gastric and Intestinal Fluid in-vitro
9 mL of generated stomach fluid as simulated in-vitro GIT juices (i.e., NaCl 125 mM, 7 mM KCl, NaHCO3 45 mM, pepsin (3 g/L, Sigma-Aldrich, USA), and pH 2.5) has been mixed with 1 mL of each strain containing 8 log CFU/mL of bacteria. Afterward, the solution was centrifuged at 6000 rpm for 10 min. Besides, the pellet was resuspended in simulated intestinal fluid containing 0.1% w/v pancreatin (Sigma-Aldrich), 0.15% w/v bile salt, and NAOH 1M to adjusted pH 8.0. Then, it was incubated at 37°C for 3 hours (22).
2.2.3. Screening of cell surface hydrophobicity
The pellet was rinsed repeatedly in PBS (pH=6.5) after centrifugation at 7,500 rpm for 15 minutes. Then, the isolates were resuspended in PBS buffer until reaching an absorption of 0.6–0.7 at 600 nm (OD0). Also, 1 mL of n-hexadecane (Merck, Germany) was mixed with 3 mL of each resuspended sample. Then, it was incubated for 30 minutes at room temperature. Afterward, the aqueous phase absorbance was determined (OD) (27).
Hydrophobicity% = (OD0 – OD/ OD0) ×100
2.2.4. Analysis of auto-aggregation and co-aggregation
The number of 8 log CFU/mL microbes has been obtained. Each strain’s solution was cultured at 37° C for various durations (i.e., 1, 2, 3, 4, and 5 hours) (23). The proportion of auto-aggregation was calculated using the following equation:
Auto-aggregation% = [(A0 – A1)/A1] ×100
where A1 and A0 denote the absorbance at times t and t = 0, respectively.
LAB isolates and pathogenic microorganisms (i.e., Escherichia coli ATCC 25922) were cultivated overnight for co-aggregation, and solutions were made as previously reported. In this case, the OD600nm was set to 0.2–0.3. Afterward, 2 mL of LAB strain was combined with 2 mL of each pathogen for 10 seconds before incubation at 37°C for 5 hours (23). The following formula has determined the co-aggregation:
Co-aggregation% = (1) ×100
Where Am expresses the combined suspension's absorbance, isolates and pathogenic bacteria have different absorbances. Al and Ap, respectively, represent these.
2.2.5. Adherence Capacity
The HT-29 cells were grown (In a CO2 incubator, BINDER INC, USA) in Dulbecco's Modified Eagle's (DMEM, Sigma-Aldrich) broth enriched with 10% fetal bovine serum (Sigma-Aldrich) and 1% penicillin-streptomycin (Sigma-Aldrich) according to standard protocole. Additionally, each strain's bacterial suspension (in DMEM medium) with 8 log CFU/mL was made, and then it was placed in the wells and incubated at 37°C for 1 hour. Also, the HT-29 cells and LAB isolates were separated (Sigma-Aldrich) with 100 μL Triton-X100. After a 10-minute incubation at 37°C, MRS broth was introduced and piped (23). The bacterial attachment percentage was estimated as follows:
Adhesion% = (adhered bacteria/Initial number of bacteria) ×100
2.3. Antimicrobial Activity
LAB strains were grown in MRS broth at 37°C for 24 hours. Then, they were centrifuged at 7500 rpm for 15 minutes, and the supernatant was sterilized by filtration (0.22µm, AmitisGen, Iran). Afterward, the supernatants were neutralized with 5N NaOH (Merck, Germany) until the pH was 6.5. Pathogens (Escherichia coli ATCC25922, Pseudomonas aeruginosa ATCC27853, Salmonella Typhimurium ATCC14028, and Staphylococcus aureus ATCC25923) were cultivated overnight in MRS agar (Conda, Spain) with 6 mm diameter wells pierced within each plate. The inhibitory zone was determined after introducing 100 μL of prepared supernatants (1×108 according to ISO19459) into wells and storing them at 37° C for 24 hours (28, 29).
2.4. Safety Analysis
2.4.1. Hemolytic Activity
LAB isolates were tested to evaluate hemolytic activity (29-31). This procedure has been performed by inoculating fresh overnight cultures on Columbia agar plates (Oxoid) containing 7% (v/v) sheep blood (Oxoid) and incubating at 37° C for 48 hours. Then, three hemolytic activities were identified (30).
2.4.2. Antibiotic Susceptibility
The antibiotic susceptibilities of LAB strains were evaluated with Tetracycline, kanamycin, chloramphenicol (e.g., 30 mg per disc), and erythromycin (e.g., 15 mg per disc). According to the Clinical and Laboratory Standards Institute tables (CLSI, 2015), isolates are classified as resistant (R), intermediate (I), or sensitive (S) (31).
2.5. Statistical Analysis
The significance of the data was determined using SPSS 19 software (SPSS Inc. Chicago, IL., USA) and Tukey's test and one-way ANOVA at a significance level of 0 .05. All studies were performed in triplicate, and the results were presented as means with standard deviations.
3.1. Isolation and Characterization
Biochemical identification of the isolated bacteria was carried out using Bergey’s manual of determinative bacteriology. Strains with Gram-positive and catalase-negative reactions were selected for further identification. Identification of LAB isolates was performed as shown (Table 1).
Table 1. The results of isolation and identification of bacteria based on physiological characteristics.
| Characteristic | Strains | ||||||||||||||||||||
| D1 | D2 | D3 | D4 | D7 | D11 | D12 | D14 | D17 | D19 | D20 | D21 | D24 | D26 | D27 | D29 | D30 | D32 | D33 | |||
| Gram staining Catalase Oxidase Indole Gas from glucose |
+ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |||
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |||
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |||
| + | + | - | + | + | + | + | + | - | + | + | + | + | + | + | + | D | + | + | |||
| Growth at temperature (ºC) | 15(ºC) | - | - | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| 45(ºC) | + | + | - | - | - | - | - | D | - | - | D | + | - | D | - | D | - | D | - | ||
| NACL | 2% | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| 4% | - | + | + | + | + | + | - | + | + | - | - | + | + | - | + | + | + | + | - | ||
| Motile Acetate from glucose Dextran from sucrose NH3 from arginine |
- | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | ||
| + | + | + | D | + | + | + | + | D | + | + | + | + | + | + | + | - | + | + | |||
| - | + | - | - | - | + | + | - | - | - | + | - | - | - | - | + | + | - | - | |||
| + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||
| Carbohydrates fermented | |||||||||||||||||||||
| Arabinose | D | + | + | + | + | - | + | + | - | - | + | + | - | D | + | + | + | + | + | ||
| Cellobiose | D | - | - | D | D | + | - | D | + | + | - | D | + | D | D | - | D | - | D | ||
| Melezitose | - | - | - | + | + | + | - | + | + | + | + | + | + | - | + | + | + | + | + | ||
| Melibiose | + | - | + | + | + | - | + | + | - | - | - | - | - | + | + | - | + | - | + | ||
| Raffinose | + | - | D | + | + | - | D | + | - | - | - | - | - | + | + | - | + | - | + | ||
| Ribose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| Sucrose | + | - | D | + | + | + | D | + | + | + | D | + | + | + | + | D | + | D | + | ||
| Xylose | D | - | + | D | D | - | + | D | - | - | - | - | - | D | D | - | D | - | D | ||
D= Positive reaction (86%)
+: Positive reaction
-: Negative reaction
Table 2 provides the molecular identification results for isolated strains. The sequencing analysis demonstrated that LAB. It belonged to seven groups, including Lactiplantibacillus plantarum, Levilactobacillus brevis, Lacticaseibacillus rhamnosus, Lacticaseibacillus paracasei, Lacticaseibacillus casei, Limosilactobacillus fermentum, and Lactobacillus jensenii.
Table 2. Molecular identification of LAB isolates using sequencing and registration of relevant genes in the NCBI database.
| LAB species | Isolate | strain species | GenBank accession no. |
| Lactiplantibacillus plantarum | D4 | Lactiplantibacillus plantarum strain 3359 | OP457175.1 |
| D7 | Lactiplantibacillus plantarum strain R LRO-7 | OR053946.1 | |
| D33 | Lactiplantibacillus plantarum strain AS.6 | OP457178.1 | |
| D27 | Lactiplantibacillus plantarum strain TPI-D27 | OR053956.1 | |
| D14 | Lactiplantibacillus plantarum strain 126X | OP558599.1 | |
| D30 | Lactiplantibacillus plantarum strain TW14-1 | OP558782.1 | |
| Lacticaseibacillus casei | D11 | Lacticaseibacillus casei strain ZBA-D11 | OR053947.1 |
| D19 | Lacticaseibacillus casei strain NCIM2151 | OP471612.1 | |
| D24 | Lacticaseibacillus casei strain SBCK12 | OP471610.1 | |
| D17 | Lacticaseibacillus casei strain 104 | OP471416.1 | |
| Lacticaseibacillus paracasei | D20 | Lacticaseibacillus paracasei strain DBR-D20 | OR053957.1 |
| D29 | Lacticaseibacillus paracasei strain WU2503 | OP467560.1 | |
| D32 | Lacticaseibacillus paracasei strain BUM12 | OP466811.1 | |
| D26 | Lacticaseibacillus paracasei strain HBUAS64030 | OP466363.1 | |
| Levilactobacillus brevis | D3 | Levilactobacillus brevis strain TMPC 46Q1A | OP464931.1 |
| D12 | Levilactobacillus brevis strain NKN55 | OP458832.1 | |
| Limosilactobacillus fermentum | D1 | Limosilactobacillus fermentum strain 4793 | OP458829.1 |
| Lacticaseibacillus rhamnosus | D21 | Lacticaseibacillus rhamnosus strain IDC-D21 | OR054188.1 |
3.2. The Beneficial Characteristics of LAB Isolates
3.2.1. Bile and Acid Tolerance
The LAB's ability to survive in the gut at a low pH is critical for surviving the early acid shock. 5 of 19 strains (i.e., L. plantarum strain TPI-D27, L. rhamnosus strain IDC-D21, L. paracasei strain DBR-D20, L. casei strain ZBA-D11, and L. plantarum strain LRO-7) were resistant to acid pH 3 (resistance rate of 50%) (Figure. 1A).
The resistance of LAB strains to pH 2, pH 3, and simulated gastric juice was done according to Table 3, and the results show that the L. rhamnosus strain IDC-D21 has the highest growth rate (6.72±0.43).
Table 3. The resistance of LAB strains to pH 2, pH 3, and simulated gastric (log CFU/mL).
| Strains/ Time | pH 3 | pH 2 | GIT juices | ||||||
| 0 hr | 1 hr | 2 hr | 3 hr | 0 hr | 1 hr | 2 hr | 3 hr | ||
| L. plantarum strain LRO-D7 | 8.05± 0.28 | 5.13±0.48 | 4.22±0.31 | 4.08±0.63 | 8.15± 0.12 | 4.67±0.42 | 3.36±0.45 | 2.01±0.37 | 4.80 ± 0.13 |
| L. casei strain ZBA-D11 | 8.18± 0.69 | 5.76±0.14 | 4.71±0.43 | 4.67±0.24 | 8.03± 0.36 | 4.32±0.46 | 3.89±0.37 | 2.94±0.56 | 4.20 ± 0.27 |
| L. paracasei strain DBR-D20 | 8.17± 0.32 | 5.61±0.28 | 4.99±0.14 | 4.45±0.55 | 8.14± 0.22 | 4.78±0.96 | 3.98±0.83 | 2.02±0.63 | 5.15 ±0.29 |
| L. rhamnosus strain IDC-D21** | 8.19± 0.14 | 8.04±0.15 | 7.35±0.48 | 6.72±0.43 | 8.09± 0.18 | 5.10±0.31 | 4.98±0.83 | 4.02±0.63 | 6.40 ± 0.25 |
| L. plantarum strain TPI-D27 | 8.13± 0.21 | 6.02±0.65 | 5.22±0.67 | 4.21±0.67 | 8.03± 0.35 | 4.12±0.36 | 3.10±0.31 | 2.76±0.34 | 4.10 ± 0.20 |
| L. casei ATCC 39392 | 8.14± 0.12 | 5.43±0.41 | 4.21±0.31 | 4.17±0.62 | 8.14± 0.16 | 4.51±0.41 | 3.31±0.40 | 2.11±0.35 | 4.70 ± 0.13 |
Revealed that only five isolates (i.e., L. plantarum strain LRO-7, L. casei strain ZBA-D11, L. rhamnosus strain IDC-D21, L. plantarum strain TPI-D27, and D30) were resistant at 0.3% (w/v) of bile salt, indicating a 50% resistance (Figure. 1B).
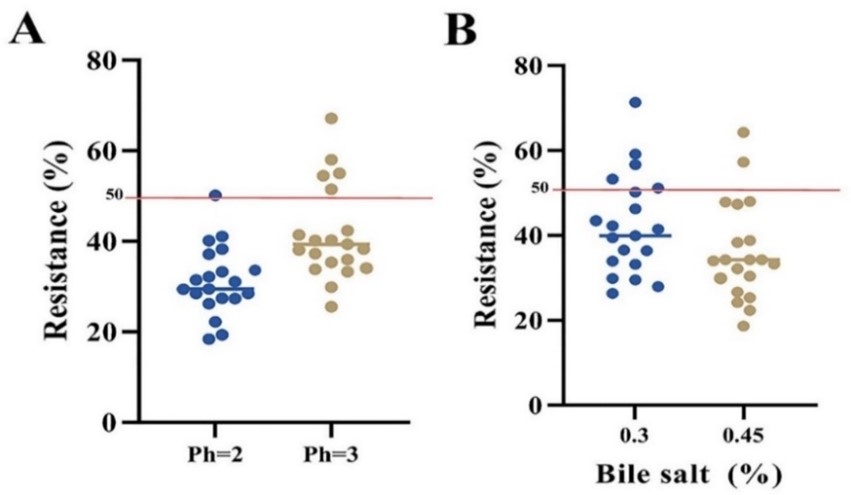
Figure 1. Resistance percentage of different bacterial strains to acidic pH (A) and bile salt (B) conditions. Isolates with at least a 50% growth rate were identified as probiotic strains.
3.2.2. Tolerance to the GIT Conditions
All isolates identified in this study (i.e., L. plantarum strain LRO-7, L. casei strain ZBA-D11, L. paracasei strain DBR-D20, and L. plantarum strain TPI-D27 showed relative resistance to these circumstances (>50%). The L. rhamnosus strain IDC-D21 had the most resistance to GIT conditions, growing 6.40 ± 0.25 after 3 hours (Table 3).
3.2.3. Cell surface hydrophobicity
The results are shown in (Figure 2), and it can be seen that The L. rhamnosus IDC-D21 and L. plantarum LRO-7 strains have the highest and lowest hydrophobicity (34.8 and 25.6%), respectively.
3.2.4. Auto-aggregation and Co-aggregation Ability
Figure 3A and 3B depict the findings of LAB strains' auto-aggregation and co-aggregation ratios.After 5 hours of incubation, the L. rhamnosus strain IDC-D21 had the highest degree of auto-aggregation (17.29%), followed by the L. paracasei strain DBR-D20 (14.28%). Also, L. plantarum strain LRO-7 had the minimum amount of auto-aggregation (9.68%) after 5 hours. All LAB strains could Co-aggregate with pathogen bacteria (Escherichia coli ATCC 25922) (Figure 3B).
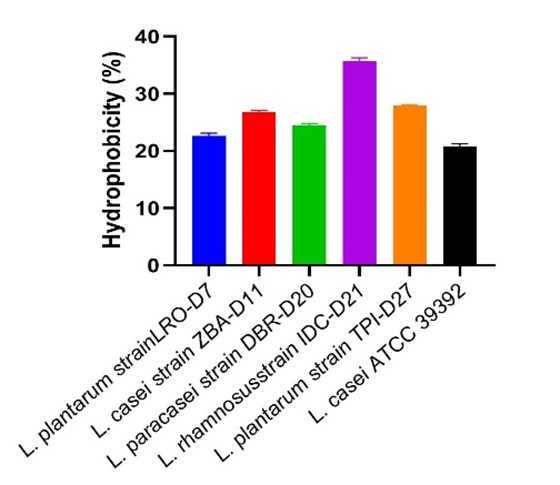
Figure 2. Cell surface hydrophobicity of LAB strains.
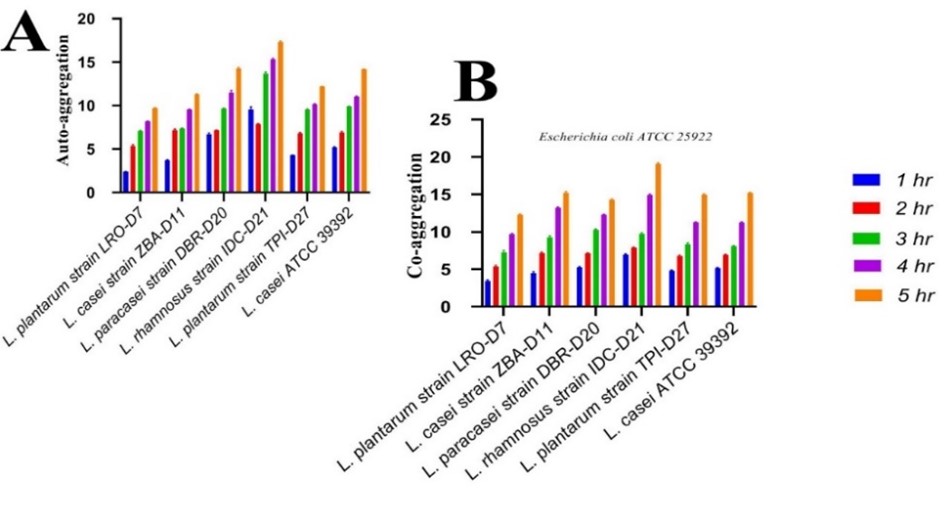
Figure 3. Examining the probiotic characteristics of the isolates. (A) Auto-aggregation percentage of LAB strains as measured at 1, 2, 3, 4, and 5 hr of incubation at 37℃. (B) Co-aggregation percentages of LAB strains with E. coli were measured after 1, 2, 3, 4, and 5 hr of incubation at 37℃ (n=3).
3.2.5. Adhesion Capacity to HT-29 Cells
All strains were able to bind to HT-29 cells to varying degrees. Bacterial adhesion percentages ranged from 7.6 to 15.4%. Also, the L. rhamnosus strain IDC-D21 and L. plantarum strain LRO-7 strains had the maximum and minimum values, respectively (Figure. 4).
3.3. Antimicrobial Activity
Table 4 presents the antimicrobial activity of LAB culture supernatant against pathogenic strains. All strains were able to inhibit pathogenic bacteria with an inhibition diameter of 7-25 mm. L. rhamnosus strain IDC-D21 was the most effective strain in inhibiting the growth of the evaluated pathogens and showed an inhibition zone of 15±0.08 to 25±0.2 mm.
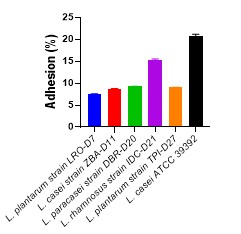
Figure 4. Examining the enzymatic activity of probiotic bacteria. LAB strain adhesion to the HT-29 cell line ( n=3)
Table 4. Antimicrobial property of LAB strains neutral pH supernatants versus pathogenic organisms.
| Strains | E. coli ATCC25922 | P. aeruginosa ATCC27853 | S.typhimurium ATCC14028 | S. aureus ATCC25923 |
| L. plantarum strain LRO-D7 | 17±0.5 | 12±0.08 | 13±0.1 | 16±0.03 |
| L. casei strain ZBA-D11 | 13±0.8 | 7±0.05 | 9±0.07 | 15±0.09 |
| L. paracasei strain DBR-D20 | 17±0.04 | 14±0.1 | 12±0.06 | 18±0.3 |
| L. rhamnosus strain IDC-D21 | 21±0.5 | 15±0.08 | 16±0.2 | 25±0.2 |
| L. plantarum strain TPI-D27 | 16±0.08 | 12±0.2 | 13±0.04 | 15±0.09 |
| L. casei ATCC 39392 | 15.7 ±0.48 | 13±0.9 | 11±0.9 | 18.01±0.24 |
| Mean±SD | 16.8 ±0.384 | 12±102 | 12.6±0.094 | 17.8±0.264 |
3.4. Safety analysis
All strains showed no hemolysis activity. The antibiotic susceptibility of Lactobacillus strains test results is reported in Table 5. In addition, L. paracasei strain DBR-D20 and L. plantarum strain TPI-D27 were resistant to two antibiotics, whereas L. rhamnosus strain IDC-D21 and L. casei strain ZBA-D11 were resistant to one antibiotic.
Table 5. Antibiotic susceptibility of LAB strains
| Strains | isolates numbers | Chloramphenicol (mm) | Erythromycin (mm) | Tetracycline (mm) | Kanamycin (mm) |
| L. plantarum strain LRO-D7 | D7 | S (22) | S (23) | I (13) | S (18) |
| L. casei strain ZBA-D11 | D11 | S (21) | S (26) | I (12) | I (15) |
| L. paracasei strain DBR-D20 | D20 | S (18) | S (29) | R (6) | R (9) |
| L. rhamnosus strain IDC-D21** | D21 | I (14) | I (19) | S (16) | R (7) |
| L. plantarum strain TPI-D27 | D27 | S (19) | S (25) | I (13) | R (10) |
| Detection threshold (CLSI) | R≤12 I 13-17 S≥18 |
R≤13 I 14-22 S≥23 |
R≤11 I 12-14 S≥15 |
R≤13 I 14-17 S≥18 |
|
S: sensitive I: intermediate R: resistant
The results obtained in this study add information to the data available in the literature regarding LAB behavior under stress conditions. L. rhamnosus strain IDC-D21 showed the highest resistance level at acid pH 3. ARAÚJO (32) that Lactiplantibacillus plantarum BJ0021 was resistant at pH 3. Also, they reported that Limosilactobacillus fermentum, Lactobacillus gasseri, and Lactobacillus delbrueckii subsp. Bulgaricus isolates had greater acid tolerance than the Lactobacillus isolated from human gastrointestinal tracts. The remaining barriers the probiotic strain must overcome to survive and multiply consist of the upper gastrointestinal system containing a stomach with a low pH and the small intestine containing bile salts and digestive enzymes (33, 34). At 0.3 and 0.45% of bile salt, isolate L. rhamnosus strain IDC-D21 exhibited the highest resistance levels. According to Gilliland (35), Lactobacilli obtained from animal intestines demonstrated resistance to bile salt higher than isolates from milk products. Patel, and Pandiella (36) provided similar results in this area. Gilliland, Staley, and Bush (37) stated that Lactobacillus acidophilus isolates obtained from calf intestinal contents grew in vitro in the presence of bile salts with substantial diversity. The chosen LAB isolates with resistance to 4% bile salts were reported by Garriga, and Pascual (38). Also, the bile salt hydrolase (BSH), an enzyme that decreases toxic effects by conjugating bile salts, accounts for the variation in resistance among lactobacilli (25). The acid pH resilience results in the present study were consistent with previous research. Acid-resistant isolates were defined as those that showed 50% resistance at an acid pH value of 3 (34, 39, 40).
Hydrophobicity and auto-aggregation are phenotypic traits. They are directly linked to strain adhesion capability. This study found that this property ranges from 14.8 to 57.3%.
In addition, co-aggregation may allow microorganisms to establish a barrier that inhibits harmful bacteria from colonizing and forming biofilms on intestinal cells and mucosal surfaces. However, auto-aggregation helps microorganisms attach to intestinal cells and mucosal surfaces (41, 42). According to Jena et al., auto-aggregation of Lactobacillus strains was isolated from rat fecal microbiota, ranging between 33.2% and 47.2%. Nevertheless, co-aggregation with pathogenic bacteria ranged from 11.89 to 38.22% at the greatest co-aggregation rate of S. aureus (43, 44).
Probiotic strains must reach intestinal cells in sufficient amounts and be alive to have a good impact. Besides, all strains could attach to HT-29 cells to varying degrees. Few studies demonstrated that biopolymers and lipoteichoic acids found on the cell membranes of Lactobacillus strains included sticky molecules that boosted their sticking potential (45, 46).
In the present study, LAB strains identified from Chaharmahal and Bakhtiari Province's indigenous raw milk yogurt have undergone in vitro testing, making them suitable candidates for probiotic and technical uses. The findings demonstrated that these strains offered high probiotic and technological possibilities. Also, the safety tests revealed that these strains were safe to eat. As a result, in vivo trials are necessary to assess their performance in real-world scenarios.
We would like to express our gratitude and thanks to all those who helped us during this research.
Ethics Approval
The Ethics Committee of the Department of Biology, College of Science, University of Sulaimai, Iraq, approved the research on 19 September 2023 (protocol code UoS-Sci-Bio 0010).
Conflicts of Interest
There is no conflict between the authors of the article.
This article was done without organizational financial support.
Received: 2023/07/17 | Accepted: 2023/09/9 | ePublished: 2024/01/29
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |








