BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-2100-en.html
2- Department of Microbiology, School of Medicine, Hamadan University of Medical Sciences, Hamadan, Iran
3- Zoonoses Research Center, Research Institute for Health Development, Kurdistan University of Medical Sciences, Sanandaj, Iran
4- Department of Microbiology, Faculty of Medicine, Kurdistan University of Medical Sciences, Sanandaj, Iran ,
Pseudomonas aeruginosa (P. aeruginosa) is a non-fermentative, gram-negative, aerobic bacillus with minimal nutritional requirements (1). It is a significant cause of a wide range of nosocomial infections, from simple to severe life-threatening infections such as urinary tract infections (UTIs), pneumonia, sepsis, bacteremia, infections in immunocompromised patients, and wound infections (1, 2). This opportunistic pathogen can also affect cystic fibrosis and burn wounds (1) and contaminate catheters, ventilators, shunts, and some disinfectants (3). These life-threatening infections are associated with high mortality and morbidity rates (4).
Treating P. aeruginosa is challenging for healthcare centers due to its intrinsic resistance to many antibiotics (4). Carbapenems (and other β-lactams) have a structure similar to acylated D-alanyl-D-alanine (the terminal amino acid residues of peptidoglycan). These antibiotics bind irreversibly to penicillin-binding proteins (PBPs), inhibiting peptidoglycan transpeptidation and disrupting cell wall synthesis (5). Carbapenem resistance in P. aeruginosa falls into three major categories: porin-mediated resistance, efflux pumps, and carbapenemases (5).
Antibiotic resistance in P. aeruginosa can be acquired through chromosomal changes or gene acquisition (6). In 2017, the World Health Organization (WHO) released a list of priority infectious bacteria, and carbapenem-resistant P. aeruginosa was classified as a high priority due to its increased mortality rate among patients with bloodstream infections (Figure 1) (7). Pseudomonas aeruginosa occupied the second rank in carbapenem-resistance (81.7±6.3%) (Figure 2) (7). Carbapenem resistance is a suitable marker for multidrug-resistant (MDR), extensively drug-resistant (XDR), and pandrug-resistant (PDR) bacteria because it indicates co-resistance to unrelated antibiotic classes (4). Determining the antibiotic resistance pattern, especially carbapenem resistance in P. aeruginosa, is a high priority for deciding effective antibiotic treatment (8).
This study analyzed the resistance patterns of imipenem and meropenem in P. aeruginosa isolates from patients referred to Kowsar Hospital in Sanandaj (year 2022) using WHONET 2022 software. The World Health Organization (WHO) released this software and allows users to modify antibiotic panels, breakpoints, and interpretations based on gender, sections, source, microbial agents (bacteria, viruses, etc.), and other data. Anyone can easily download this software and use it offline.
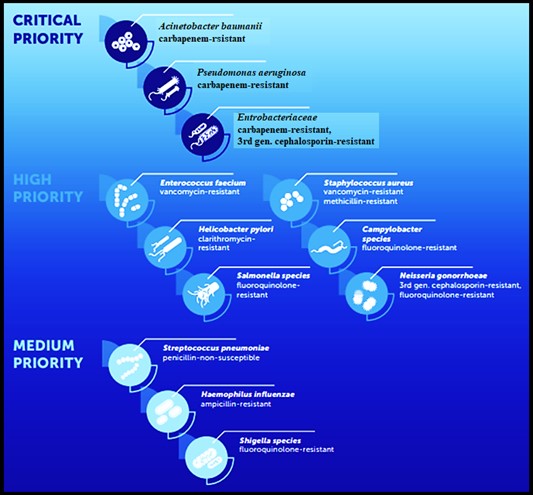
Figure 1. Priority pathogens for antibiotics (6)
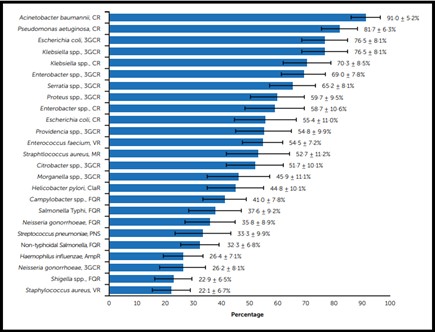
Figure 2. Ranking of antibiotic-resistant bacteria (mean weight and standard deviation) AmpR: ampicillin-resistant, CR: carbapenem-resistant, ClaR: clarithromycin-resistant, FQR: fluoroquinolone-resistant, MR: methicillin-resistant, PNS: penicillin non-susceptible 3GCR: third-generation cephalosporin-resistant, VR: vancomycin-resistant (6).
Specimen Collection
During one year (February 2022-February 2023), 95 P. aeruginosa isolates were collected from patients referred to Kowsar Hospital (a teaching hospital in Sanandaj, west of Iran). Specimens were obtained from various sites such as urine, blood, shunts, ascites, bronchoalveolar lavage (BAL), and sinuses. Isolates were identified by growth on a selective medium (Cetrimide agar), biochemical tests such as oxidase, citrate, O/F test, urease test, triple sugar iron (TSI), and colony characteristics like pigmentation and odor (9). For final identification, the 16sRNA gene PCR (135 bp) was used (forward primer -CTTACGGCCAGGGCTACACA- and reverse primer -GCGATTCCGACTTCACGCAG-). PCR was carried out with 10ng template DNA, 0.24 μM of each primer, and Master mix (sinaclon.Co.) in a total volume of 25 μL. The amplification protocol included an initial denaturation (94˚C for 3 minutes) followed by 35 cycles of denaturation (94˚C for 60 seconds), annealing (62˚C for 30 seconds), and extension (72˚C for 60 seconds), with a single final extension of 10 minutes at 72˚C. The PCR product was detected on a 1% agarose gel for 45 minutes at 100 volts. The positive control in this study was P. aeruginosa ATCC27853 (9).
Antimicrobial Susceptibility Testing
According to the Clinical and Laboratory Standards Institute (CLSI 2021) protocol, antibacterial susceptibility tests were performed for imipenem (IPM 10µg, Padtan Teb Co.) and meropenem (MEM 10µg, Padtan Teb Co.) using the disk diffusion method (10). After incubation for 24 hours at 37˚C, P. aeruginosa isolates were classified as susceptible, intermediate, or resistant. P. aeruginosa ATCC27853 was used as the control strain (10).
Data Analysis
Interpreted results were imported into WHONET 2022 software as data. There were no duplicate strains and only the first isolate from each patient was used.
In this study, 95 P. aeruginosa isolates were examined. Of the 95 isolates, 44 (46.3%) and 51 (53.7%) were isolated from women and men, respectively. These isolates were recovered from various sites, with the highest frequencies belonging to tracheal, urine, and blood samples at 30 (31.6%), 22 (23.2%), and 21 (22%), respectively (Table 1). Figure 3 shows the PCR results and band patterns.
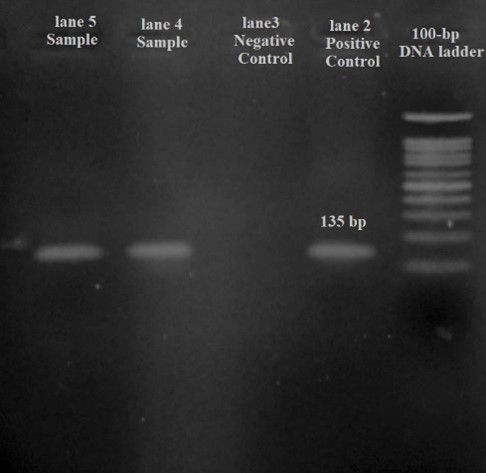
Figure 3. Polymerase chain reaction (PCR) assay for P.aeruginosa detection. Lane 1: 100-bp DNA ladder (SinaClon, Tehran, Iran); lane 2: PCR-positive control (135 bp); Lanes 3: negative control Lane 4-5: positive PCR products.
Table 1. Frequency of P. aeruginosa isolation sites
| Samples | Tracheal | Urine | Blood | Wound | Ascites | BAL | Sinus |
| Frequency (%) | 30 (31.6%) | 22 (23.2%) | 21 (22%) | 18 (19%) | 2 (2%) | 1 (1.1%) | 1 (1.1%) |
Antibiotic Resistance: Of the 95 isolates, 83 (87.4%) and 61 (64.2%) were resistant to IPM and MEM, respectively (Table 2). Interpretation with WHONET 2022 software showed that there is one major route for resistance to imipenem and meropenem in P. aeruginosa isolates. Still, other ways exist that were used by these isolates to become resistant to imipenem and meropenem (Figures 4 and 5). In total, simultaneous resistance to imipenem and meropenem was observed in 59 (62.1%) of the isolates (Figure 6).
Table 2. Frequency of IPM and MPN susceptibility test for P. aeruginosa isolates
| Susceptibility Antibiotic |
S | I | R |
| Imipenem (IPM) | 2 (2.1%) | 10 (10.5%) | 83 (87.4%) |
| Meropenem (MEM) | 31 (32.7%) | 3 (3.1%) | 61 (64.2%) |
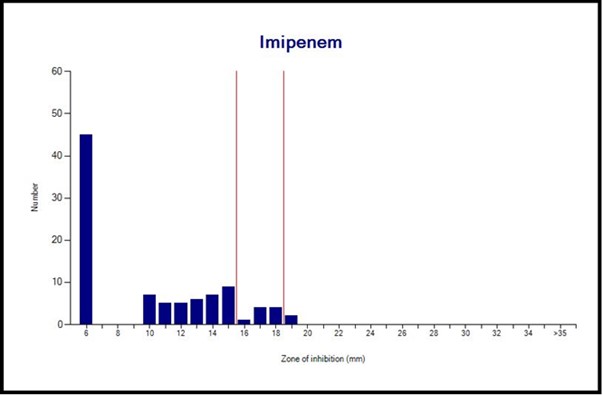
Figure 4. Imipenem susceptibility pattern of P. aeruginosa isolates
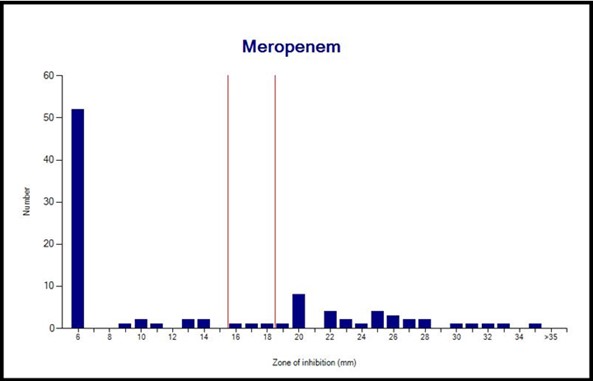
Figure 5. Meropenem susceptibility pattern of P. aeruginosa isolates
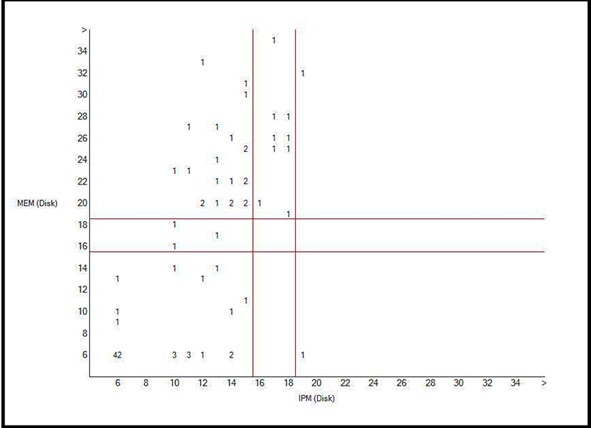
Figure 6. Scatter Plot of imipenem and meropenem resistance P. aeruginosa isolates
Table 3. P. aeruginosa resistance patterns for imipenem and meropenem in different provinces of Iran (8).
| Province | IPM (%) | MEN (%) |
| Tehran | 70.4 | 78.8 |
| Isfahan | 76.1 | 93 |
| Orumieh | 30.8 | 39.4 |
| Guilan | 23.3 | NA |
| Zahedan | 17.2 | NA |
| Ahvaz | 42.9 | 44.1 |
| Zanjan | 29.2 | NA |
| Hamedan | 7.5 | 13.2 |
| Tabriz | 49 | NA |
| Total | 31.6 | 40 |
| NA: Not available | ||
In the present study, the frequency of P. aeruginosa resistance to IPM and MEM was 83 (87.4%) and 61 (64.2%), respectively. A study on antibiotic resistance patterns of P. aeruginosa from 2010 to 2017 in different provinces of Iran showed a range from 7% to 76.1% and from 13.2% to 93% for IPM and MEM, respectively (Table 3) (8). In another study in Iran from 2020 to 2021, out of 115 P. aeruginosa isolates, 52.2% were resistant to carbapenems (9). A study in Turkey showed that out of 58 P. aeruginosa isolates, 57 (99%) were resistant to IPM and 43 (74%) were resistant to MEM (11). A retrospective study on 128 P. aeruginosa isolates during 2017-2022 using WHONET software (3) found that out of 124 isolates, 22% were resistant to carbapenems. Incomplete information in this study may have caused differences in our results. A study on resistance patterns of 3036 P. aeruginosa isolates during five years (2017-2022) in Guangdong Province (China) using WHONET software showed that resistance rates for imipenem and meropenem were 22.4% and 19.3%, respectively (12). One major reason for the differences between our study and recent studies could be the COVID-19 outbreak. At that time, antibiotic prescriptions increased and led to the emergence of strains with greater antibiotic resistance. Additionally, during the COVID-19 pandemic, some people took antibiotics arbitrarily without a doctor's prescription, which increased antibiotic resistance.
In a study conducted in Delhi in 2019, data analysis was done with WHONET software, and the resistance pattern of several bacteria, including P. aeruginosa, was obtained from data provided by 24 participating laboratories. The results showed that 44% and 42.9% of P. aeruginosa isolates were resistant to imipenem and meropenem, respectively (13). As mentioned earlier, during the year when the COVID-19 pandemic began, due to the high consumption of antibiotics, the chance of the appearance of antibiotic-resistant strains increased. Therefore, during epidemic and pandemic events, antibiotics must be prescribed based on antibiotic stewardship.
Overall, our findings are within the range of most P. aeruginosa resistance patterns for IPM and MEM reported in studies conducted in Iran and other countries. According to previous experiences, the resistance of Pseudomonas aeruginosa to IPM and MEM could be related to one or more resistance mechanisms to these antibiotics. In our study, analysis with WHONET software showed that there was one main mechanism of resistance and some P. aeruginosa isolates used different mechanisms for resistance to IPM and MEM. The mechanisms for carbapenem resistance are porin-mediated resistance, efflux pumps, and carbapenemases (5). WHONET is a useful software that can assist not only in managing data in clinical microbiology laboratories but also as a tool for monitoring and comparing antimicrobial susceptibility and resistance patterns over a long period.
This software is useful for monitoring clinical interventions, controlling nosocomial infection programs, making management decisions, and formulating hospital policies. Additionally, WHONET software can be a suitable method for preliminary analysis of theses and studies. It is recommended to use this software to create a network in cities or provinces to monitor antibiotic resistance.
We would like to thank the Kurdistan University of Medical Sciences, the Research Deputy of Kurdistan University of Medical Sciences, and the Cellular & Molecular Research Center. We also thank Kowsar Hospital in Sanandaj for providing specimens and for data gathering.
Conflicts of Interest
This study does not include any conflicts of interest for the authors.
None.
Received: 2023/06/19 | Accepted: 2023/09/21 | ePublished: 2023/11/29
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |








