BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-2069-en.html
2- Department of Medical Laboratory Techniques, Nasiriyah Technical Institute, Southern Technical University, Nasiriyah, Iraq ,
3- Department of Nursing Techniques, Nasiriyah Technical Institute, Southern Technical University, Nasiriyah, Iraq
Intestinal helminth Enterobius vermicularis (E. vermicularis) is a nematode transmitted by ingesting the eggs found in water, food, fomites, and dust. Enterobiasis affects more than 40 million people in the USA and is associated with families of low socioeconomic status (1). E. vermicularis is the only nematode from the family Oxyurida that infects humans and causes enterobiasis. Rapid transmission occurs due to the absence of an intermediate host (2). The average lifespan of this nematode is 2-3 months, a period necessary to complete its life cycle in the host (3). Adult females mature within one month, while males die after fertilizing female worms. Gravid females migrate to the large intestine, laying eggs (about 2,000 per day) on the perianal skin (4). They reside in the large intestine but may also be found in the ectopic locations such as the appendix, uterus, kidneys, urinary tract, eyes, female genital tract, and subcutaneous nodules (5).
Enterobiasis can be asymptomatic or associated with pruritus, abdominal pain, insomnia, restlessness, irritability, enuresis, and in some cases, secondary infection of the scratched perianal skin (6). Misdiagnosis due to the lack of reliable diagnostic tests can lead to acute appendicitis. The sticky tape test and direct smear examination have low sensitivity, resulting in many undiagnosed cases and making enterobiasis control difficult (7, 8). Polymerase Chain Reaction (PCR) is a highly sensitive molecular diagnostic technique that relies on the isolation of nematodes or eggs, which can be challenging to obtain from patients with enterobiasis (9). DNA extraction and purification from stool samples are difficult due to the consistency of feces and the presence of many PCR inhibitors (10-12). Extracting and purifying DNA with adequate methodology is necessary, especially when using PCR for E. vermicularis characterization (13, 14). The internal transcribed spacer 2 (ITS2) region is a good genetic marker for identification and phylogenetic studies, and ITS2 rDNA sequences are valuable for exploring inter- and intra-population genetic differences in parasites (15).
The current study was designed to determine the molecular identification of E. vermicularis in Nasiriyah city, Thi-Qar Province, and to detect genetic variation in the small subunit of ribosomal RNA (ITS2) gene.
Study Population and Sample Collection
Stool samples were collected using clean, dry, and sterile containers from 56 infected patients, who attended to Bint Al-Huda Pediatric Teaching Hospital at Al-Nasiriyah City in Thi-Qar province, south of Iraq. All samples were confirmed with microscopic examination. Sixty apparently healthy people were selected as control group. All samples were stored by freezing at -20°C. The participants were given verbal consent for their participation in this study.
Stool DNA Extraction
DNA samples from the stool specimens were isolated using Presto™ Stool DNA Extraction Kit. Briefly, the specimens (200 mg) were added into the bead-beating tubes containing ceramic beads and ST1 lysis buffer (800 μL) was added. The tubes were incubated at 70°C for 5 min and centrifuged at 8,000 x g for 2 min, and then 500 μL of the supernatant was transferred into a different 1.5 ml microcentrifuge tube. The ST3 buffer (800 μL) was added to the tube. Then, 700 μL stool sample was transferred to the GD column in the collection tube and centrifuged at 16,000 x g for 1 min. The GD column was transferred to another collection tube. The last step was repeated and centrifuged at 16,000 x g for 30 sec. The column was washed with Wash buffer (600 μL) and centrifuged at 16,000 x g for 30 sec. Then, preheated elution buffer (100 μL) was added into the center of the column matrix and centrifuged at 16,000 x g for 2 min for DNA elution and purification. The isolated DNA sample's quality and quantity were tested using a Nanodrop spectrophotometer (THERMO, USA) at 260/280 nm. Free nuclease water was used as blank.
Primer Design
The primers for detection of E. vermicularis included internal transcribed spacer 2 (ITS2) in ribosomal DNA gene. The Primer 3 plus and NCBI Gene Bank sequence database (HQ646164.1) were used for primer design. The primers were manufactured by Scientific Researcher Co. Ltd, Iraq as forward sequence: 5’-GCTGCTGCGGTTAAAAAGCT-3’, and 5’-reverse Sequence: AGCAGGTTTGAGTCTCGCTC-3’ with product size 750 bp.
PCR Reaction
GoTaq ®Green PCR Master Mix kit was used for the preparation of PCR master mix with a total volume of 20 µL composed of 5 µL DNA template, 2 µL of each primer (10 pmol), 12.5 µL GoTaq ®Green PCR Master Mix, and 3.5 µL PCR water. The component of the master mix was placed into an Exispin vortex centrifuge at 3000 x g for 3 min, then located in a conventional PCR Thermocycler system. The cycling conditions were as follows: Initial denaturation (95°C for 5 min), then 35 cycles of denaturation (95°C for 30 s), annealing (58°C for 30 s), and extension (72°C for 2 min), and then the final extension (72°C for 5 min). The PCR products were evaluated by running on agarose gel electrophoresis. The 100 bp Molecular Ladder was added in parallel. The PCR products were visualized using UV Transilluminator (Accurise Instruments Transcat Company, USA).
DNA Sequencing Analysis
DNA sequencing was done to determine the genetic association and analyze the genetic variation in the rDNA ITS2 region in local E. vermicularis isolates. The DNA sequencing was also applied as a confirmative identification for the PCR-positive human E. vermicularis worms and to study phylogenetic association by analyzing the phylogenetic tree. The phylogenetic tree was constructed using Unweighted Pair Group method with Arithmetic Mean (UPGMA tree) in MEGA software version 6.0.
Only 32 (57.14%) of 56 samples were confirmed positive by PCR. The analysis of ITS2 in the ribosomal DNA gene of E. vermicularis isolates was confirmed, as shown in Figure 1.

Figure 1. The ITS2 PCR products run on agarose gel. M: marker (100-1500 bp), lanes 1-10: positive E. vermicularis isolates (750 bp).
The method of DNA sequencing was accomplished for ITS2 gene in local E. vermicularis isolates for PCR positive human E. vermicularis, and the homology sequence identity results between local E. vermicularis IQN.1-10 isolates and NCBI-BLAST E. vermicularis from South Korea and Germany databases exhibited a genetic homology ranged from 99.54- to 99.56% as showed in Table 1.
Table 1. The identity of NCBI-BLAST homology sequence percentage between local E. vermicularis IQN.1-IQN.10 isolates and NCBI-BLAST closely related E. vermicularis South Korea and Germany isolates.
| Local E. vermicularis isolate |
Accession number | Homology sequence identity (%) | ||
| Country related NCBI | Accession number | Identity (%) | ||
| IQN.1 | OM100902.1 | South Korea and Germany | KU680848.1-MN914074.1 | 99.55% |
| IQN.2 | OM100903.1 | South Korea and Germany | KU680848.1-MN914074.1 | 99.55% |
| IQN.3 | OM100904.1 | South Korea and Germany | KU680848.1-MN914074.1 | 99.56% |
| IQN.4 | OM100905.1 | South Korea and Germany | KU680848.1-MN914074.1 | 99.55% |
| IQN.5 | OM100906.1 | South Korea and Germany | KU680848.1-MN914074.1 | 99.56% |
| IQN.6 | OM100907.1 | South Korea and Germany | KU680848.1-MN914074.1 | 99.54% |
| IQN.7 | OM100908.1 | South Korea and Germany | KU680848.1-MN914074.1 | 99.56% |
| IQN.8 | OM100909.1 | South Korea and Germany | KU680848.1-MN914074.1 | 99.55% |
| IQN.9 | OM100910.1 | South Korea and Germany | KU680848.1-MN914074.1 | 99.55% |
| IQN.10 | OM100911.1 | South Korea and Germany | KU680848.1-MN914074.1 | 99.55% |
The genetic analysis of the ITS2 region revealed low diversity in the local E. vermicularis IQN.1-10 isolates. The genetic variations or mutations analysis of the ITS2 gene sequences showed that the substitution and deletion mutations were very low, and only three deletion and substitution mutations were found at total genetic variation percentages ranging from 0.44 to 0.46%, as shown in Table 2. Finally, the local E. vermicularis IQN.1-10 isolates were submitted into NCBI GeneBank by accession numbers from OM100902.1 into OM100911.1.
Table 2. The NCBI-BLAST genetic variation analysis between local E.vermicularis IQN.1-IQN.10 isolates and NCBI-BLAST closely related E. vermicularis South Korea and Germany isolates.
| Local E. vermicularis isolate |
Accession number | Homology sequence identity (%) | ||
| Number of Mutations | Type of Mutation | Mutation % | ||
| IQN.1 | OM100902.1 | 3 | Adenine, thymine, Guanine | 0.45% |
| IQN.2 | OM100903.1 | 3 | Adenine, thymine, thymine | 0.45% |
| IQN.3 | OM100904.1 | 3 | Adenine, thymine, cytosine | 0.44% |
| IQN.4 | OM100905.1 | 3 | Adenine, thymine, Guanine | 0.45% |
| IQN.5 | OM100906.1 | 3 | Adenine, thymine, Guanine | 0.45% |
| IQN.6 | OM100907.1 | 3 | Adenine, thymine, Guanine | 0.46% |
| IQN.7 | OM100908.1 | 3 | Adenine, thymine, Guanine | 0.44% |
| IQN.8 | OM100909.1 | 3 | Adenine, thymine, cytosine | 0.45% |
| IQN.9 | OM100910.1 | 3 | Adenine, thymine, cytosine | 0.45% |
| IQN.10 | OM100911.1 | 3 | Adenine, thymine, cytosine | 0.45% |
OM100905.1_IQN.4 ---CCGTTTAGCTGGTTCTCCTAGCGTTGCCTT-TACCGGTCGCGTTAGG
OM100904.1_IQN.3 ---CCGTTTAGCTGGTTCTCCTAGCGTTGCCTT-TACCGGTCGCGTTAGG
OM100903.1_IQN.2 ---CCGTTTAGCTGGTTCTCCTAGCGTTGCCTT-TACCGGTCGCGTTAGG
OM100911.1_IQN.10 ---CCGTTTAGCTGGTTCTCCTAGCGTTGCCTT-TACCGGTCGCGTTAGG
OM100909.1_IQN.8 ---CCGTTTAGCTGGTTCTCCTAGCGTTGCCTT-TACCGGTCGCGTTAGG
OM100910.1_IQN.9 ---CCGTTTAGCTGGTTCTCCTAGCGTTGCCTT-TACCGGTCGCGTTAGG
OM100906.1_IQN.5 ---CCGTTTAGCTGGTTCTCCTAGCGTTGCCTT-TACCGGTCGCGTTAGG
OM100902.1_IQN.1 ---CCGTTTAGCTGGTTCTCCTAGCGTTGCCTT-TACCGGTCGCGTTAGG
OM100907.1_IQN.6 ---CCGTTTAGCTGGTTCTCCTAGCGTTGCCTT-TACCGGTCGCGTTAGG
OM100908.1_IQN.7 ---CCGTTTAGCTGGTTCTCCTAGCGTTGCCTT-TACCGGTCGCGTTAGG
JF934731.1_South.Korea ---CCGTTTAGCTGGTTCTCCTAGCGTTGCCTT-TACCGGTCGCGTTAGG
MT260072.1_Congo ---CCGTTTAGCTGGTTCTCCTAGCGTTGCCTT-TACCGGTCGCGTTAGG
MN914074.1_Germany ---CCGTTTAGCTGGTTCTCCTAGCGTTGCCTT-TACCGGTCGCGTTAGG
FR687850.1_Czech ACTCTGAATAGCTATAGTTGATCGCATGGTCTTGTACCGGCGGC-----G
AB626601.1_Japan ACTCTGAATAGCTATAGTTGATCGCATGGTCTTGTACCGGCGGC-----G
* * ***** * * ** * * *** ****** ** *
OM100905.1_IQN.4 TGGCTAGCAAGT--TTACTTTGAAAAAATTAGAGTGCTTAAAGCGGGCTA
OM100904.1_IQN.3 TGGCTAGCAAGT--TTACTTTGAAAAAATTAGAGTGCTTAAAGCGGGCTA
OM100903.1_IQN.2 TGGCTAGCAAGT--TTACTTTGAAAAAATTAGAGTGCTTAAAGCGGGCTA
OM100911.1_IQN.10 TGGCTAGCAAGT--TTACTTTGAAAAAATTAGAGTGCTTAAAGCGGGCTA
OM100909.1_IQN.8 TGGCTAGCAAGT--TTACTTTGAAAAAATTAGAGTGCTTAAAGCGGGCTA
OM100910.1_IQN.9 TGGCTAGCAAGT--TTACTTTGAAAAAATTAGAGTGCTTAAAGCGGGCTA
OM100906.1_IQN.5 TGGCTAGCAAGT--TTACTTTGAAAAAATTAGAGTGCTTAAAGCGGGCTA
OM100902.1_IQN.1 TGGCTAGCAAGT--TTACTTTGAAAAAATTAGAGTGCTTAAAGCGGGCTA
OM100907.1_IQN.6 TGGCTAGCAAGT--TTACTTTGAAAAAATTAGAGTGCTTAAAGCGGGCTA
OM100908.1_IQN.7 TGGCTAGCAAGT--TTACTTTGAAAAAATTAGAGTGCTTAAAGCGGGCTA
JF934731.1_South.Korea TGGCTAGCAAGT--TTACTTTGAAAAAATTAGAGTGCTTAAAGCGGGCTA
MT260072.1_Congo TGGCTAGCAAGT--TTACTTTGAAAAAATTAGAGTGCTTAAAGCGGGCTA
MN914074.1_Germany TGGCTAGCAAGT--TTACTTTGAAAAAATTAGAGTGCTTAAAGCGGGCTA
FR687850.1_Czech TGTCTATCAAGTGTCTGCCTTATCAACTTTCGATGGTAGTTTATGTGCCT
AB626601.1_Japan TGTCTATCAAGTGTCTGCCTTATCAACTTTCGATGGTAGTTTATGTGCCT
** *** ***** * * ** ** ** ** * * **
OM100905.1_IQN.4 TCGATGCTTGAATAGTGGTGCATGGGATAATAGAATACGATTACGGGTCT
OM100904.1_IQN.3 TCGATGCTTGAATAGTGGTGCATGGGATAATAGAATACGATTACGGGTCT
OM100903.1_IQN.2 TCGATGCTTGAATAGTGGTGCATGGGATAATAGAATACGATTACGGGTCT
OM100911.1_IQN.10 TCGATGCTTGAATAGTGGTGCATGGGATAATAGAATACGATTACGGGTCT
OM100909.1_IQN.8 TCGATGCTTGAATAGTGGTGCATGGGATAATAGAATACGATTACGGGTCT
OM100910.1_IQN.9 TCGATGCTTGAATAGTGGTGCATGGGATAATAGAATACGATTACGGGTCT
OM100906.1_IQN.5 TCGATGCTTGAATAGTGGTGCATGGGATAATAGAATACGATTACGGGTCT
OM100902.1_IQN.1 TCGATGCTTGAATAGTGGTGCATGGGATAATAGAATACGATTACGGGTCT
OM100907.1_IQN.6 TCGATGCTTGAATAGTGGTGCATGGGATAATAGAATACGATTACGGGTCT
OM100908.1_IQN.7 TCGATGCTTGAATAGTGGTGCATGGGATAATAGAATACGATTACGGGTCT
JF934731.1_South.Korea TCGATGCTTGAATAGTGGTGCATGGGATAATAGAATACGATTACGGGTCT
MT260072.1_Congo TTGATGCTTGAATAGTGGTGCATGGAATAATAGAATACGATTACGGATCT
MN914074.1_Germany TCGATGCTTGAATAGTGGTGCATGGGATAATAGAATACGATTACGGGTCT
FR687850.1_Czech ACCATGGTTG--TTACGGGTAACGGAGAATTAGGGTTCGACTCCGG--AG
AB626601.1_Japan ACCATGGTTG--TTACGGGTAACGGAGAATTAGGGTTCGACTCCGG--TG
*** *** * ** * ** * *** * *** * ***
OM100905.1_IQN.4 ATTTTGTTGGTTTTCTGATCTGTGATAATGGTTAAGAGGGACAAACGGGG
OM100904.1_IQN.3 ATTTTGTTGGTTTTCTGATCTGTGATAATGGTTAAGAGGGACAAACGGGG
OM100903.1_IQN.2 ATTTTGTTGGTTTTCTGATCTGTGATAATGGTTAAGAGGGACAAACGGGG
OM100911.1_IQN.10 ATTTTGTTGGTTTTCTGATCTGTGATAATGGTTAAGAGGGACAAACGGGG
OM100909.1_IQN.8 ATTTTGTTGGTTTTCTGATCTGTGATAATGGTTAAGAGGGACAAACGGGG
OM100910.1_IQN.9 ATTTTGTTGGTTTTCTGATCTGTGATAATGGTTAAGAGGGACAAACGGGG
OM100906.1_IQN.5 ATTTTGTTGGTTTTCTGATCTGTGATAATGGTTAAGAGGGACAAACGGGG
OM100902.1_IQN.1 ATTTTGTTGGTTTTCTGATCTGTGATAATGGTTAAGAGGGACAAACGGGG
OM100907.1_IQN.6 ATTTTGTTGGTTTTCTGATCTGTGATAATGGTTAAGAGGGACAAACGGGG
OM100908.1_IQN.7 ATTTTGTTGGTTTTCTGATCTGTGATAATGGTTAAGAGGGACAAACGGGG
JF934731.1_South.Korea ATTTTGTTGGTTTTCTGATCTGTGATCATGGTTAAGAGGGACAAACGGGG
MT260072.1_Congo ATTTTGTTGGTTTTCTGATCTGTGACAATGGTTAAGAGGGACAAACGGGG
MN914074.1_Germany ATTTTGTTGGTTTTCTGATCTGTGATAATGGTTAAGAGGGACAAACGGGG
FR687850.1_Czech AGGGAGCTTGAGAAATGG-CTACCACATCCAAGGAAGGCAGCAGGCGCGC
AB626601.1_Japan AGGGAGCTTGAGAAATGG-CTACCACATCCAAGGAAGGCAGCAGGCGCGC
* * * * ** ** * * * ** ** *
OM100905.1_IQN.4 GCATTCGTATCGCTGCGCGAGAGGTGAAATTCGTGGACCGTAGCGAGACG
OM100904.1_IQN.3 GCATTCGTATCGCTGCGCGAGAGGTGAAATTCGTGGACCGTAGCGAGACG
OM100903.1_IQN.2 GCATTCGTATCGCTGCGCGAGAGGTGAAATTCGTGGACCGTAGCGAGACG
OM100911.1_IQN.10 GCATTCGTATCGCTGCGCGAGAGGTGAAATTCGTGGACCGTAGCGAGACG
OM100909.1_IQN.8 GCATTCGTATCGCTGCGCGAGAGGTGAAATTCGTGGACCGTAGCGAGACG
OM100910.1_IQN.9 GCATTCGTATCGCTGCGCGAGAGGTGAAATTCGTGGACCGTAGCGAGACG
OM100906.1_IQN.5 GCATTCGTATCGCTGCGCGAGAGGTGAAATTCGTGGACCGTAGCGAGACG
OM100902.1_IQN.1 GCATTCGTATCGCTGCGCGAGAGGTGAAATTCGTGGACCGTAGCGAGACG
OM100907.1_IQN.6 GCATTCGTATCGCTGCGCGAGAGGTGAAATTCGTGGACCGTAGCGAGACG
OM100908.1_IQN.7 GCATTCGTATCGCTGCGCGAGAGGTGAAATTCGTGGACCGTAGCGAGACG
JF934731.1_South.Korea GCATTCGTATCGCTGCGCGAGAGGTGAAATTCGTGGACCGTAGCGAGACG
MT260072.1_Congo GCATTCGTATCGCTGCGCGAGAGGTGAAATTCGTGGACCGTAGCGAGACG
MN914074.1_Germany GCATTCGTATCGCTGCGCGAGAGGTGAAATTCGTGGACCGTAGCGAGACG
FR687850.1_Czech AAATTACCCACTCTCGGTATGAGGAGGTAGTGACGAGAAATAACGAGACC
AB626601.1_Japan AAATTACCCACTCTCGGTATGAGGAGGTAGTGACGAGAAATAACGAGACC
*** * ** * **** * * * * ** ******
OM100905.1_IQN.4 CCCTACTGCGAAAGCATTTGCCAAGAATGTTTTCATTAATCAAGAACGAA
OM100904.1_IQN.3 CCCTACTGCGAAAGCATTTGCCAAGAATGTTTTCATTAATCAAGAACGAA
OM100903.1_IQN.2 CCCTACTGCGAAAGCATTTGCCAAGAATGTTTTCATTAATCAAGAACGAA
OM100911.1_IQN.10 CCCTACTGCGAAAGCATTTGCCAAGAATGTTTTCATTAATCAAGAACGAA
OM100909.1_IQN.8 CCCTACTGCGAAAGCATTTGCCAAGAATGTTTTCATTAATCAAGAACGAA
OM100910.1_IQN.9 CCCTACTGCGAAAGCATTTGCCAAGAATGTTTTCATTAATCAAGAACGAA
OM100906.1_IQN.5 CCCTACTGCGAAAGCATTTGCCAAGAATGTTTTCATTAATCAAGAGCGAA
OM100902.1_IQN.1 CCCTACTGCGAAAGCATTTGCCAAGAATGTTTTCATTAATCAAGAGCGAA
OM100907.1_IQN.6 CCCTACTGCGAAAGCATTTGCCAAGAATGTTTTCATTAATCAAGAGCGAA
OM100908.1_IQN.7 CCCTACTGCGAAAGCATTTGCCAAGAATGTTTTCATTAATCAAGAACGAA
JF934731.1_South.Korea CCCTACTGCGAAAGCATTTGCCAAGAATGTTTTCATTAATCAAGAACGAA
MT260072.1_Congo CCCTACTGCGAAAGCATTTGCCAAGAATGTTTTCATTAATCAAGAAC---
MN914074.1_Germany CCCTACTGCGAAAGCATTTGCCAAGAATGTTTTCATTAATCAAGAACGAA
FR687850.1_Czech GTTCTCATTGAGGCCGGTTATCG-GAATGAGTTG---GATCTAAATGTCC
AB626601.1_Japan GTTCTCATTGAGGCCGGTTATCG-GAATGAGTTG---GATCTAAATGTCC
* ** * ** * ***** ** *** * *
Figure 2. Analysis of multiple sequence alignment for the small subunit ribosomal RNA gene in the local E. vermicularis IQN.1-IQN.10 isolates with related isolates from other countries in NCBI GenBank. Analysis of multiple alignments was created using the Online ClustalW alignment tool. Alignment analysis showed the nucleotide alignment similarity as (*) and substitution mutations of the small subunit rRNA gene between isolates.
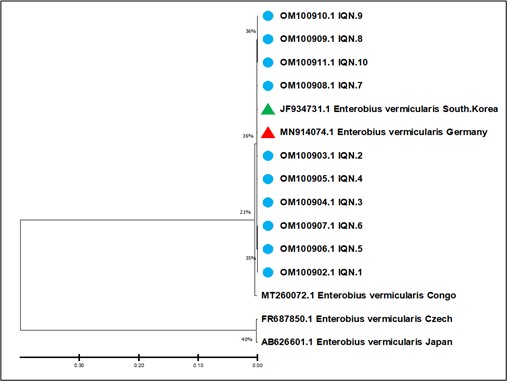
Figure 3. Analysis of phylogenetic tree was based on the small subunit rRNA gene partial sequence in E. vermicularis IQN.1-IQN.10 isolates that were used for the genetic association analysis. The Un-weighted Pair Group method created the phylogenetic tree with Arithmetic Mean (UPGMA tree) in MEGA version 6.0. The E. vermicularis IQN.1-IQN.10 isolates revealed a close relationship to the NCBI-BLAST E. vermicularis South Korea and Germany isolates at total genetic changes of 0.03-0.010%.
Molecular techniques have so far contributed to enhancing diagnosis. PCR is a very sensitive technique, more so than other traditional diagnostic laboratory techniques. It is used to diagnose many pathogenic agents, and recent studies suggest that environmental PCR testing can be very valuable in assessing and managing pathogenic agent outbreaks (16).
In a recent study, DNA was isolated and purified from the fecal samples. The isolation process was difficult due to the small amount of nematode DNA in the samples and the presence of the PCR inhibitors in the stool. This agrees with the study of Schielke (17) in Germany, which showed that the presence of inhibitors in a large range of sample types can lead to a decreased sensitivity of PCR or even false-negative PCR results. The fecal samples were chosen for this study because they were obtained from patients. This was in agreement with the study of Ummarino, Caputo (9) in Italy that developed a PCR diagnostic method to diagnose E. vermicularis in the stool samples. Molecular studies of this nematode can increase diagnostic sensitivity. It also provides knowledge about the diversity, evolutionary relationships with the host, and geographical distribution (18, 19).
Our sequencing analysis results for the ITS2 region showed high similarity and very low numbers of mutations. This high similarity may be due to high conserved ITS2 region of the ribosomal DNA. This agrees with the results of Tomanakan and Sanpool (20) in Thailand, which found high similarity between ITS2 sequences of their E. vermicularis isolates. It also agrees with the results of Zelck Ulrike, Bialek and Weiß (21) in Germany, where no ribosomal DNA diversity was found between E. vermicularis isolates and children. These results disagree with Le, Blair and McManus (22) study in Australia based on the mitochondrial DNA sequences proving high genetic variability in many helminth groups.
Our study phylogenetic analysis was done based on the ITS2 gene. A few studies have been performed on this gene marker, while many other studies were based on high-variable gene sequences like the mitochondrial COX1 gene, which is a preferred marker for phylogenetic analysis in the study of Piperaki, Spanakos (23) in Greece, Shafiei, Jafarzadeh (24), and Kaneva, Harizanov (25) in Bulgaria. This may explain the homology of the isolates in these studies with the isolates from Denmark, Greece, Germany, and Japan (13, 26, 27). The genetic analysis of local E. vermicularis IQN.1-10 isolates showed fewer correlations to NCBI-Blast E. vermicularis isolates from other countries, probably due to the regional genetic variation; this outcome agrees with the study of Kadhum, Jaber and Alkhanaq (28) in Iraq that revealed the IQ-K1-5 isolates were out of the tree and less correlated to the NCBI-Blast E. vermicularis isolates. Our study results also agree with the results of Janthu, Dumidae (29) study in Thailand that showed E. vermicularis from humans had a low diversity of ITS2 region.
This study confirms the importance and the efficacy of molecular techniques, mainly PCR, to improve the diagnosis of Enterobius vermicularis and understand its genetic diversity, evolutionary relationships, and geographic distribution. The ITS2 ribosomal DNA region proved to be a valuable marker for the diagnostic and epidemiological implications due to its high conservation and high sequence similarity and low mutation rates observed in our isolates. The results of this study indicate a new understanding of the E. vermicularis genotypes in Iraq, emphasizing further molecular studies that are to be conducted across different regions for a better understanding of the genetic landscape for this parasitic nematode.
We acknowledge the Bint Al-Huda Hospital laboratory technicians for their help with sample collection. We extend our thanks to the Public Health Department in the Health Office of Thi-Qar Province for their support. We also appreciate the Thi-Qar Education Directorate, Nasiriyah, Iraq, the Department of Medical Laboratory Techniques, and the Department of Nursing Techniques at the Nasiriyah Technical Institute, Southern Technical University, Nasiriyah, Iraq, for providing the necessary resources and facilities.
Ethical Considerations
The protocol for this study was approved by the Public Health Department management/Thi-Qar Health Office to collect and examine the stool samples (Ref.No.28-06.04.2022).
Authors’ Contributions
Conceptualization: Manar Karem Kadhim. Data curtain: Mohammed Hassan Flaih. Formal analysis: Ruaa Majid Khazaa. Methodology: Manar Karem Kadhim and Mohammed Hassan Flaih. Project administration: Ruaa Majid Khazaa and Khwam Reissan Hussein. Visualization: Ruaa Majid Khazaa. Writing of the original draft: Manar Karem Kadhim and Khwam Reissan Hussein. Writing – review & editing: Mohammed Hassan Flaih, Ruaa Majid Khazaa, Khwam Reissan Hussein, and Manar Karem Kadhim.
This research received no specific grant from any funding agency in the public.
Conflicts of Interest
The authors have no conflicts of interest to declare.
Received: 2024/04/27 | Accepted: 2024/08/1 | ePublished: 2024/08/18
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |








