BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1860-en.html
2- Department of Pharmaceutical Microbiology, Faculty of Pharmacy, Olabisi Onabanjo University, Ago Iwoye, Ogun State, Nigeria
3- Department of Medical Laboratory Science, College of Medicine, University of Lagos, Lagos State, Nigeria
4- Department of Microbiology, Faculty of Life Sciences, Modibbo Adama University, Yola, Adamawa State, Nigeria
Staphylococcus is a prominent genus that has been implicated in opportunistic infections in both humans and animals (1). The genus is classified into two main groups, coagulase positive and negative Staphylococcus. Coagulase is an enzyme and one of the basic virulent factors found in Staphylococcus aureus and other species of Staphylococci. The enzyme is responsible for plasma clotting and it is used to identify S. aureus (2). S. aureus can cause gastroenteritis due to the production of enterotoxins. This bacterium is regarded as one of the most frequent food-borne pathogens (3). They have been recognized as pathogenic Gram-positive bacteria of public health concerns. The family is made up of ten genera. Recently, the genus Mammaliicoccus was taken out of the family and assigned with five species previously under Staphylococcus. The type species of the newly created genus is M. sciuri (4). The reclassification of the five species was based on phylogenomic analyses due to the perception that the five were heterotypic synonyms. Hence, the five species were taxonomically reassigned to the new genus Mammaliicoccus gen. nov (4) Members of the genus Staphylococcus and Mammaliicoccus are normal microbial flora of the skin and mucosa parts of humans and animals as well as soil. Staphylococcal infections could affect everywhere including organs in the human body, leading to surface and deep abscesses, respiratory as well as urinary tract infections (5). Staphylococci and Mammaliicocci species have been regarded as a group of bacteria with a tendency to cause debilitating infections when the opportunity arises (6). Most of these infections are associated with methicillin and multidrug resistance (7).
Antimicrobial resistance (AMR) has become a worldwide threat to human and animal health, with tedious and expensive treatment and possible mortality (3). Not long after the clinical application of methicillin as an antibiotic-resistant, it emerged, as a result of methicillin-resistant gene mecA, found on Staphylococcal cassette chromosome mec (SCCmec), a genetic mobile element (8, 9). The global occurrence of methicillin-resistant Staphylococci has become a threat to public health. This could be due to the ease with which genes for resistance are acquired among the family members of Staphylococcaceae. The formation of biofilm by the strains of S. aureus has been mentioned to be an important factor in the development of infections as well as resistance to methicillin by S. aureus (10) The interaction between animals, wild or domesticated, and man could be another factor for the spread of the resistance genes (11).
Methicillin-resistant M. sciuri has been reported in ruminants and New World camelids, harboring antimicrobial-resistant genes that are found in other closely related and unrelated species (12). Mammaliicoccus species are thought to be involved in the transmission of resistant genes and serve as reservoirs for these genes found among the members of the family Staphylococcaceae, especially the well-known S. aureus. The abuse of antibiotics to treat microbial infections in human and animal has been mentioned to increase antibiotic resistance among the Staphylococci. Methicillin-resistant Staphylococci are known to be resistant to virtually all classes of antibiotics. When a bacterium is resistant to at least one member of three or more classes of antibiotics, such a bacterium is said to be multi-drug resistant (2, 13, 14). The report of Bardasheva et al. (15) showed that coagulase-positive and negative S. haemolyticus and S. epidermidis were both methicillin and multidrug-resistant.
The success of antibiotic treatment of methicillin-resistant Staphylococcus is relatively dependent on promptness to avoid nosocomial infections. Strains of S. haemolyticus are known to cause infections in some hospital patients such as those on dialysis, diabetics, and post-surgery patients as well as the development of resistance to many antibiotics in bedridden individuals (16, 17). This study was carried out to isolate and characterize methicillin and multidrug-resistant Staphylococci isolated from wounds of patients with cases of wound infection at Federal Medical Center, Yola, Adamawa State, Nigeria, and the possible presence of some virulent indicators.
Samples Collection and Isolation of Staphylococci Species
Forty-five clinical samples were collected from patients using sterile cotton swabs within Federal Medical Centre, Yola, Adamawa State, Nigeria. The samples were transported on ice packs to the General Laboratory, Department of Biotechnology, Modibbo Adama University, Yola, Adamawa State, Nigeria. Isolation was done by streaking on Mannitol Salt Agar. The plates were incubated at 37˚C for 18 to 24 h aerobically. Individual colonies suspected to be Staphylococci were sub-cultured on freshly prepared Mannitol Salt Agar (MSA) and incubated at 37˚C for 18 – 24 h. The pure cultures of the isolates were recovered and sub-cultured on nutrient agar slant and incubated at 37˚C for 18 – 24 h. These pure culture slants were kept in the refrigerator (Haier Thermocool) at 4˚C until they were needed for other analyses.
Screening for Methicillin and Multi-Drug Resistant Staphylococcal Species
Suspected Staphylococci isolates were screened for methicillin resistance using the disk diffusion method as described by Hudzicki (18). Staphylococcal isolates that were positive for methicillin resistance were further screened for multi-drug resistance using a Gram-positive multi-drug antibiotic disc (Biomark Laboratories, India). Both methicillin and multi-drug resistant screening were carried out on Mueller Hinton Agar. Cefoxitin antibiotic disc (Rapid Lab, UK) was used to screen for methicillin-resistant while antibiotic disc comprising Vancomycin (VAN) 30 µg, Ceftazidime (CPZ) 10 µg, Cephalexin (CP) 1.5 µg, Ampicillin (AMP) 10 µg, Meropenem (MEM) 10 µg, Erythromycin (ERY) 5 µg, Tetracycline (TET) 30 µg, Cotrimoxazole (COT) 25 µg, Cefuroxime (CRX) 10 µg, Gentamicin (GEN) 10 µg, Ciprofloxacin (CIP) 5 µg, and Augmentin (AUG) 30 µg were used for multi-drug antibiotic screening. The results were measured using a metric ruler and interpreted as either resistance (R), intermediate (I), or sensitive (S) using the CLSI standard (19).
Determination of Multiple Antibiotic Resistance Indices of the Four Staphylococci Isolates
The multiple antibiotic resistance index of each of the isolates M. sciuri HFS1, S. haemolyticus HFS2, M. sciuri HFS3, and Staphylococcus spp. HFS4 was determined by dividing the number of antibiotics each of the isolates was resistant to by the total number of antibiotics applied in the study as described by Krumperman (20).
Sugar Fermentation and Biochemical Tests
Biochemical characteristics and sugar fermentation using arabinose, xylose, inositol, mannitol, fructose, lactose, maltose, glucose, sucrose and galactose, catalase, urease, citrate, TSI, oxidase, and indole tests were carried out to have a presumptive identity of the isolates (21).
DNA Extraction
Deoxyribonucleic acid (DNA) of the four selected Staphylococci that were methicillin and multi-drug resistant were extracted using an overnight growth culture in nutrient broth. A DNA extraction kit (model 24700) manufactured by Norgen, Canada was used according to the instructions of the manufacturer.
Amplification and Partial Sequencing of 16S rDNA Gene
The 16S rDNA was amplified using the primers are ST16-F-5’-TTGCTTCTCTGATGTTAGCG-3’, ST16-R-3’- AATCATTTGTCCCACCTT-5’ (22). The mix consisted of NEB one Taq master mix with standard buffer (catalog No MO4825) 10 μL, genomic DNA 1.0 μL, forward and reverse primers each 1.0 μL and nuclease-free water (catalog No E476) 7.0 μL were prepared to run the PCR. The initial denaturation was done at 94˚C for 5 min, followed by 35 cycles of denaturation at 94˚C for 1.0 min. The annealing took place at 50˚C for 30 sec while the extension took place at 68˚C for 1.0 min and the final extension at 68˚C for 1 min.
The integrity of the PCR amplicon was visualized on a 1 % agarose gel (CCL-AG500, Cleaver Scientific Ltd). A molecular ladder of 100 bp (NEB’s fast DNA ladder) was added to serve as a standard. The bands were stained with EZ-Vision Blue light DNA Dye. The amplicon was purified by PCR clean-up and then sequenced in an automated DNA sequencer using the dideoxy chain termination method (23).
Construction of the phylogenetic tree
The sequences of the 16S rDNA gene of three out of four (one failed to amplify) selected Staphylococci were compared with similar sequences of Staphylococci strains deposited at GenBank using the Entrez search engine at the National Center for Biotechnology Information (NCBI). The similarity in sequences of the isolated Staphylococci and deposited Staphylococci strains at GenBank was done using the BLAST program. Additionally, multiple sequence alignment and phylogenetic tree construction were done with molecular evolutionary genetics analysis (MEGA X) (24).
The 16S rDNA gene sequences of three Staphylococci were deposited at GenBank with assigned accession numbers.
Amplification of Coagulase coa, Methicillin mec A, and Enterotoxigenic SEA Genes
Two genes, coa, and SEA were amplified using the respective primers coa F 5’-ATAGAGATGCTGGTACA-GG-3’, coa R 3’GCTTTCCGATTGTTCGATGC-5’ for coagulase gene, and SEA-A2-5’ATTAACCGAAAG-TTCTGTAGA-3’, SEA-U2-3’TTGCGTAAAAGTCTGAATT-5’ for the enterotoxigenic gene in the different reaction mix. The amplicon of each gene was observed on agarose gel of 1.0 % with a molecular ladder of 100 bp (NEB’s fast DNA ladder) (CCL-AG500, Cleaver Scientific Ltd) while the staining of the bands was done with EZ-Vision Blue light DNA Dye.
Statistical Analysis
Data collected in this study were analyzed using simple mean ± standard deviation (Mean±SD) and presented using bar charts.
Isolation and Identification of the Staphylococci Species
Thirty-one suspected Staphylococci isolates were recovered from wound samples collected from patients attending Federal Medical Center, Yola, Adamawa State, Nigeria. They were mannitol fermenters based on their cultural characteristics on mannitol salt agar. Four of the isolates were coagulase-positive, resistant to methicillin, and resistant to not less than five antibiotics tested that cut across all the classes of antibiotics (Table 1).
Table 1. Screening for Multi-Drug Resistant Staphylococci isolates
| Bacterial strain |
ERY (15 μg) |
TET (30 μg) |
COT (25 μg) |
CRX (10 μg) |
GEN (10 μg) |
CIP 5 μg |
AMP 10 μg |
MEM 10 |
AUG 30 μg |
CP 1.5 μg |
CPZ 10 μg |
VAN 30 μg |
| 1. M. sciuri HFS1 | 19.4±.4 | 18.9±0.6 | 16.7±0.4 | R | 16.9±0.4 | 21.9±0.6 | R | R | R | 8.1±0.3 | R | 13±0.8 |
| 2. S. haemolyticus HFS2 | R | R | R | R | 25.9±0.8 | R | R | R | R | 23.9±0.6 | R | R |
| 3. M. sciuri HFS3 | 8.9±0.6 | R | R | R | 7.1±0.8 | 7.0±0.8 | 9.9±0.4 | R | R | R | R | 15.9±0.8 |
| 4. Staphylococcus sp. HFS4 | 19.9±0.4 | 9.7±1.0 | R | 7.7±0.8 | R | 9.0±0.4 | R | R | R | R | R | 11.9±0.4 |
Key: R = Resistant, ERY = Erythromycin, TET = Tetracycline, COT = Cotrimoxazole, CRX = Cefuroxime, GEN = Gentamicin, CIP = Ciprofloxacin, AMP = Ampicillin, MEM = Meropenem, AUG = Augmentin, CP = Cephalexin, CPZ = Ceftazidime and VAN = Vancomycin, ≤ 9. 0 mm = Resistant, 10.0-13.0 mm = Intermediate and ≥ 14.0 mm = Susceptible
The four presumptive Staphylococci isolates were identified using their biochemical and molecular characteristics (Table 2, Figure 1). Biochemically, the four isolates M. sciuri HFS1, S. haemolyticus HFS2, M. sciuri HFS3 and Staphylococcus spp. HFS4 exhibited characteristics of the Staphylococci with little differences among them. They all fermented most 6-C sugars and were unable to utilize the 5-C sugars such as xylose, inositol, and arabinose (Table 2). All four isolates HFS1, HFS2, HFS3, and HFS4 were unable to ferment sucrose, inositol, and xylose while only HFS2 and HFS4 utilized arabinose and lactose as their carbon and energy sources, respectively. However, all four isolates metabolized galactose, glucose, maltose, fructose, and mannitol as their carbon and energy sources (Table 2). Isolates HFS1, HFS2, HFS3, and HFS4 were positive for catalase, and coagulase and were able to utilize citrate. All the isolates except HFS4 were positive for TSI. However, none of the isolates metabolize urea. Likewise, they were all negative in reaction to oxidase and indole tests (Table 2).
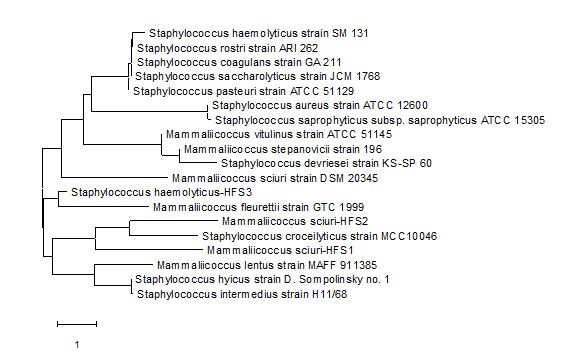
All four isolates HFS1, HFS2, HFS3, and HFS4 were identified using the sequences of the 16S rDNA gene. But only three HFS1, HFS2, and HFS3 gave satisfactory amplification that enabled them to be identified using the computer program Basic Local Alignment Search Tool (BLAST) at NCBI. Isolates HFS1 and HFS3 were identified to belong to the genus Mammaliicoccus and they were tagged Mammaliicoccus sciuri HFS1 (ON340756) and M. sciuri HFS3 (ON340770) while HFS2 was identified to be a strain of Staphylococcus haemolyticus, hence it was tagged Staphylococcus haemolyticus HFS2 (ON358435). All three sequenced isolates clustered closely with other species of Staphylococci (Figure 2).
Methicillin and Antimicrobial Resistance Screening
The four methicillin-resistant isolates had zones of inhibition that ranged between 0.0 mm to 9.0 mm with cefoxitin and they were coagulase-positive strains, S. haemolyticus HFS2, Staphylococcus spp. HFS4, M. sciuri HFS1, and M. sciuri HFS3 screened for multidrug resistance using a multidisc antibiotic (Biomark Laboratories, India) that contained twelve different antibiotics that cut across the major classes of antibiotics such as cephalosporin, fluoroquinolone, carbapenems, glycopeptides, aminoglycosides, and penicillin.
Table 2. Biochemical characteristics and sugar fermentation of the Staphylococci isolates
| Bacterial strain | Gala | Sucro | Gluco | Malt | Lact | Fruct | Mannit | Inosit | Xylo | Arabino | Cata | Coagu | Citrat | Urea | TSI | Oxi | Indole |
| 1. M. sciuri HFS1 | +ve | -ve | +ve | +ve | -ve | +ve | +ve | -ve | -ve | -ve | +ve | +ve | +ve | -ve | +ve | -ve | -ve |
| 2. S. haemolyticus HFS2 | +ve | -ve | +ve | +ve | -ve | +ve | +ve | -ve | -ve | -ve | +ve | +ve | +ve | -ve | +ve | -ve | -ve |
| 3. M. sciuri HFS3 | +ve | -ve | +ve | +ve | -ve | +ve | +ve | -ve | -ve | -ve | +ve | +ve | +ve | -ve | +ve | -ve | -ve |
| 4. Staphylococcus spp. HFS4 | +ve | -ve | +ve | +ve | +ve | +ve | +ve | -ve | -ve | -ve | +ve | +ve | +ve | -ve | -ve | -ve | -ve |
Keys: Gala = Galactose, Sucro = Sucrose, Gluco = Glucose, Malt = Maltose, Lact = Lactose, Fructo = Fructose, Mannit = Mannitol, Inosit = Inositol, Xylo = Xylose, Arabino = Arabinose, Cata = Catalase, Coagu = Coagulase, Citrat = Citrate, TSI = Triple sugar iron and Oxi = Oxidase, +ve = Positive reaction and –Ve = Negative reaction
Figure 1. Phylogenetic tree showing the taxonomic relationship of Mammaliicoccus sciuri-HFS1, M. sciuri-HFS2, and Staphylococcus haemolyticus-HFS3 with other Staphylococci isolates at GenBank

Figure 2. Agarose gel electrophoresis of PCR product of 16S rDNA gene amplicon of 1.0 kb of isolates A, B, C, and D. NEB fast ladder (M) 0.1 kb was the standard marker used
A = M. sciuri HFS1 B = S. haemolyticus HFS2 C = M. sciuri HFS3 D = Staphylococcus spp. HFS4
Table 3. Multi-drug resistance pattern of the four Staphylococci isolates
| Bacterial strain | Resistance phenotype | Class of antibiotics |
| M. sciuri HFS1 | CRX, AMP, MEM, AUG, CP, CPZ | Penicillins, Carbapenems, Cephalosporins, |
| S. haemolyticus HFS2 | ERY, TET, COT, CRX, CIP, AMP, MEM, AUG, CPZ, VAN | Macrolides, Penicillins, Sulfonamides, Cephalosporins, Carbapenems, Glycopeptides, Fluoroquinolones |
| M. sciuri HFS3 | ERY, TET, COT, CRX, GEN, CIP, MEM, AUG, CP, CPZ | Macrolides, Cephalosporins, Sulfonamides, Carbapenems, Aminoglycosides, Fluoroquinolones, |
| Staphylococcus spp. HFS4 | COT, CRX, GEN, CIP, AMP, MEM, AUG, CP, CPZ | Sulfonamides, Cephalosporins, Penicillins, Fluoroquinolones, Carbapenems, |
Key: ERY = Erythromycin, TET = Tetracycline, COT = Cotrimoxazole, CRX = Cefuroxime, GEN = Gentamicin, CIP = Ciprofloxacin, AMP = Ampicillin, MEM = Meropenem, AUG = Augmentin, CP = Cephalexin, CPZ = Ceftazidime and VAN = Vancomycin
Table 4. Multiple Antibiotics Resistance (MAR) Index of the four Staphylococci isolates
| Bacterial strain | Number of antibiotics tested (b) | Number of antibiotics resistant to (a) | MAR index (a/b) |
| M. sciuri HFS1 | 12 | 6 | 0.50 |
| S. haemolyticus HFS2 | 12 | 10 | 0.80 |
| M. sciuri HFS3 | 12 | 10 | 0.80 |
| Staphylococcus sp. HFS4 | 12 | 9 | 0.75 |
Amplification of the coagulase gene, coa was observed in S. haemolyticus HFS2 and M. sciuri HFS3, but not in the remaining two isolates, though all four strains had a positive reaction to the coagulase biochemical test (Figure 3, Table 2). None of the isolates studied had their enterotoxigenic gene SEA amplified after four attempts running the PCR, showing that they all lack this particular gene. The presence of coa gene has been a measure of virulence in S. aureus. The amplicon size of about 600 bp was generated for the coa gene in S. haemolyticus HFS2 and M. sciuri HFS3.

Figure 3. Agarose electrophoresis PCR product (cropped) for coagulase (coa) gene (600 bp) of S. haemolyticus HFS2 and Mammaliicoccus sciuri HFS3 and isolates A and D (not amplified, but biochemically positive). Lane M is NEB fast ladder
B = S. haemolyticus HFS2 C = M. sciuri HFS3
All four isolates HFS1, HFS2, HFS3, and HFS4 were unable to ferment sucrose, inositol, and xylose while only HFS2 and HFS4 utilized arabinose and lactose as their carbon and energy sources respectively. However, they were able to metabolize galactose, glucose, maltose, fructose, and mannitol as their carbon and energy sources. The reported observations in this study in sugar fermentation, enzymes catalase and coagulase production as well as oxidase and indole reaction are similar to the earlier report by Gotz et al. (5). The utilization of any carbon source and production of specific enzymes are under the influence of genes controlling the reaction and environmental factors. Similarly, all four strains were negative for indole and oxidase tests Gotz et al. (5). These two biochemical tests are used to presumptively biochemically identify Staphylococci isolates (25).
The amplicon of the 16S rDNA was found to be around 1.0 kb. The use of PCR amplification of 16S rDNA specific primers has been mentioned to identify Staphylococcal isolates up to the species and subspecies level (22), who reported an amplicon size of 1.4 kb for 16S rDNA gene of the Staphylococci isolated by them.
Methicillin and multi-drug resistant strains of Staphylococci are becoming a public health burden due to the tediousness and cost of treatment. They are responsible for nosocomial infections which have led to an increase in morbidity and mortality rates (2). Sands et al. (26) reported the isolation of methicillin-resistant strains of S. epidermidis, S. haemolyticus, and M. sciuri that are resistant to routinely prescribed antibiotics. The methicillin resistance could be due to the possession of different mec genes present in the staphylococcal cassette chromosome mec (SCCmec), a genetic mobile element (International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (9).
Another factor that has been mentioned to be responsible for the spread of methicillin and antimicrobial resistance genes is the ease with which the genes are acquired among the family members of Staphylococcaceae as well as wide distribution of species of Staphylococcus and Mammaliicoccus among human and animals (9). M. sciuri is a reservoir for the mecA gene though it may not be expressed in the strain when acquired and mutated within the promoter region, it could be expressed in S. aureus (5) Species of Mammaliicoccus have been implicated to carry genes that make them to be resistant to many antibiotics (6, 27). Bardasheva et al. (15) had previously mentioned the resistance of coagulase-positive S. haemolyticus and S. epidermidis to be methicillin and multidrug-resistant. High resistance of Staphylococcal species to ampicillin, tetracycline, ciprofloxacin, and erythromycin has been mentioned by Osada et al. (27), Marincola et al. (7), Sadiq et al. (28).
The multiple antimicrobial resistance indices for the four isolates studied ranged from 0.5 to 0.8. A similar report of high indices has been recently reported by Mir et al. (29), though with Salmonella sp., while Asante et al. (30) reported a high index of 0.8 with Staphylococci species associated with nosocomial infections. The high values of indices are indications of high-risk contamination which could be responsible for multi-drug resistant strains observed in this study. The values we report in this study are higher than 0.2, which is regarded as the standard (19). The observations we are reporting in this study are similar to a previous report by Gotz et al. (5).
Sugar Fermentation and Biochemical Characteristics of the Staphylococci
Biochemically, the four isolates M. sciuri HFS1, S. haemolyticus HFS2, M. sciuri HFS3 and Staphylococcus spp. HFS4 exhibited characteristics of the Staphylococci with little differences among them. They all fermented most 6-C sugars and were unable to utilize the 5-C sugars such as xylose, inositol arabinose, and mannitol (15). These observations are similar to the previous report by Gotz et al. (5). All four isolates were catalase and coagulase enzyme producers making them potential virulent and pathogenic strains (15, 31). Coagulase for instance is produced by Staphylococci to initiate blood plasma clotting in host plasma (31).
Amplification of Coagulase coa and Enterotoxigenic SEA Genes
Amplification of the coagulase gene, coa was seen in S. haemolyticus HFS2 and M. sciuri HFS3, but not in the remaining two isolates, though all four strains had a positive reaction to the coagulase biochemical test. The positive reaction to the coagulase enzyme test could be attributed to the presence of genes for the enzyme in these strains while the negatives recorded in the PCR amplification could be due to experimental error or nonbinding of the primers to the specific gene. None of the isolates studied had their enterotoxigenic gene SEA amplified after four attempts running the PCR, showing that they all lack this particular gene. The presence of coa gene has been a measure of virulence in S. aureus. In this study, an amplicon size of about 600 bp was generated for the coa gene in S. haemolyticus HFS2 and M. sciuri HFS3. Varied values of amplicon sizes that ranged between 681 bp to 891 bp have been earlier reported by Sadiq et al. (28). A similar study carried out in India and the United Kingdom showed that the amplicon size of the coa gene could range between 510 bp and 1000 bp when same primers were used (31).
This study showed that Staphylococci species of Staphylococcus and Mammaliicoccus genera are methicillin, multidrug-resistant, and coagulase as well as catalase positive. They are potential pathogens with multiple genes that have been reported to be resistant to most commonly available drugs in developing countries like Nigeria. All four isolates studied M. sciuri HFS1, S. haemolyticus HFS2, M. sciuri HFS3, and Staphylococcus spp. HFS4 were resistant to not less than five antibiotics that cut across most classes of antibiotics. The multiple antimicrobial indices ranged from 0.5 to 0.8. Isolation of these four strains from clinical samples is a great public health concern, as their presence indicates a high-risk area and may worsen antimicrobial resistance in immune-compromised patients. These may lead to an economic burden on treatment, high morbidity and mortality rates, and death. Therefore, there is a need for regular antimicrobial surveillance within the hospital, timely detection of methicillin and multi-drug resistant Staphylococci, and administration of a cocktail of more potent antibiotics in the treatment of wounds and other patients within the hospital environment.
All the authors sincerely appreciate the support received from the management of Federal Medical Centre, Yola, (FMC, Yola) the staff of the Medical Microbiology Department, FMC, Yola, the technical staff of the Biotechnology Department, Modibbo Adama University, Yola.
The authors jointly funded this research.
Conflicts of Interest
No conflict of interest was declared by the authors.
Received: 2023/01/15 | Accepted: 2023/06/19 | ePublished: 2023/09/27
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |