BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1751-en.html

 , Hami Kaboosi1
, Hami Kaboosi1 
 , Seyed Reza Mohebbi2
, Seyed Reza Mohebbi2 
 , Hamid Asadzadeh Aghdaei3
, Hamid Asadzadeh Aghdaei3 
 , Mohammad Reza Zali Zali4
, Mohammad Reza Zali Zali4 

2- Gastroenterology and Liver Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran ,
3- Basic and Molecular Epidemiology of Gastrointestinal Disorders Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
4- Gastroenterology and Liver Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Hepatitis B virus (HBV) infection is still a major public health problem despite the execution of vaccination programs and various therapeutic regimens. Transmission of chronic hepatitis B (CHB) occurs either at birth or through person-to-person transmission. The disease affects 350 million people worldwide (1, 2). High-risk sexual behaviors and using contaminated injecting devices are among the factors of horizontal disease transmission between individuals (3). Chronic hepatitis B infection is a highly variable course of interaction between virus and host immune responses. Therefore, it has different manifestations in individuals (4). The disease can also cause extrahepatic manifestations, including skin changes, mixed cryoglobulinemia, polyarteritis nodosa, and glomerulonephritis (5). People with hepatitis B infection are at risk for the disease to progress to fibrosis, cirrhosis, and hepatocellular carcinoma (HCC) (6). Therefore, timely treatment is essential to control this world health problem and prevention of disease progression, and the treatment process depends on an optimal diagnostic method. There are methods for diagnosing HBV infection and related diseases based on clinical parameters such as blood tests including aminotransferases (ALT, AST), HBV DNA levels, fibrosis-related factors, serological tests, and the evaluation of HBV-specific antigens and anti-HBV antibodies (7). There is also a needle liver biopsy diagnostic procedure that is difficult and invasive despite the accuracy and can put the patient at risk (8).
MicroRNAs (miRNAs) are a class of non-coding RNA molecules with approximately 22 nucleotides that negatively regulate gene expression post-transcriptionally by interacting with the mRNA of target genes (9). MiRNAs regulate various cellular pathways and processes, including differentiation, cell proliferation, angiogenesis, development, apoptosis, metabolism, and immune responses (10, 11). Studies show that miRNAs influence the expression of genes through regulatory pathways, for example, BCL2 by miR-15a, PTEN by miR21, E2F1 by miR-17, RAS by Let-7 and other genes and regulatory pathways in various diseases (12-16). Some studies demonstrate that miRNAs in serum and plasma could discriminate hepatocellular carcinoma from other liver diseases and specific differences of miRNAs expression can indicate a specific stage of liver disease (17-31). Studies have shown that miR-222 increases the differentiation and proliferation of liver stellate cells (HSCs) which downregulate the expression of the intercellular adhesion molecule 1 (ICAM-1). As the ICAM-1 is responsible for transporting immune cells to the site of liver damage, this process will increase the rate of liver damage. Therefore, in this study, we decided to analyze the possible changes of miR-222 expression patterns in patients with chronic hepatitis B.
Study Population and miRNA Selection
This study enrolled 43 patients (18 chronic active hepatitis and 25 inactive carriers, including 23 male and 20 female) and 43 healthy (24 male and 19 female) individuals referred to Taleghani Hospital, Tehran, Iran. As very few biomarker studies have been performed on the role of miRNA-222 in HBV chronic infection previously, the present study was performed as a pilot study with 43 subjects in each group. All the patients were tested for anti-HCV and anti-HDV antibodies and were negative for these serological test results. The HBsAg-positive and Anti-HBc Antibody positive individuals were selected as chronic hepatitis B patients' group (HBsAg was positive for more than 6 months) and HBsAg-negative and Anti-HBc Antibody negative individuals as a control group. Exclusion criteria were history of other liver diseases, including other viral hepatitis C and D, HIV infection, history of hepatocellular carcinoma and other cancers, and pregnancy for female participants. A general practitioner examined all the patients and controls, and then patients were interviewed and rechecked by a GI medical specialist. Ethics Committee approved the study of the Research Institute for Gastroenterology and liver diseases, Shahid Beheshti University of Medical Sciences (IR.SBMU.RIGLD.REC.1395.97). Informed consent was obtained from all individual participants included in the study. Healthy individuals had normal liver function and no history of liver diseases. The diagnosis of CHB in the patient group was determined according to their laboratory, clinical and ultrasound results. Non-parametric Mann-Whitney test was used to compare the mean age of patients and controls, which showed a significant difference (P=0.539). The miRNAs data based were used to select proper option with various targeted genes utilizing TargetScan (http://www.targetscan.org) database (release 8.0) and miRWalk (http://mirwalk.umm.uni-heidelberg.de/) software (version 3). NCBI's Gene Expression Omnibus (GEO) Data Sets with GEO Series access numbers GSE69580 and GSE49012 (https://www.ncbi.nlm.nih.gov/gds/?term=GSE69580+GSE49012) were also checked to evaluate the selected miRNA. OligoAnalyzer Tool (www.idtdna.com) and the miRBase database also utilized for the primer design process.
Laboratory and Biochemical Measurements
Serum levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST), were determined by a standard automated biochemistry analyzer (912; Hitachi Co., Q., Japan).
Plasma miRNA Preparation
Participants' blood samples were collected in EDTA tubes. The samples were centrifuged at 3,000 rpm for 15 minutes at 4°C. The resultant plasma samples were stored at -80°C for downstream processes. According to the manufacturer's protocol, RNA extraction was performed by the miRNeasy Serum/Plasma Kit (Qiagen, Germany). Then for cDNA synthesis, we used the Mir-X miRNA First-Strand Synthesis Kit (Takara/Clontech, CA, US). In the Mir-X cDNA synthesis reaction, the RNAs are poly (A)-tailed by using poly (A) polymerase and copied using a modified oligo (dT) primer and SMART MMLV reverse transcriptase. For relative quantification of miR-222, we used qRT-PCR SYBR Premix Ex Taq II (Tli RNaseH Plus, Takara Japan) according to the manufacturer's instructions with a dedicated primer for miR-222 (Table 1). MiRNA-222-3p specific 5' primer with oligo sequence comprising of 5'- AGCTACATCTGGCTACTGG -3' and mRQ 3' Primer supplied with the Mir-X™ miRNA First-Strand Synthesis Kit were utilized to quantify the amplicon. Master mix including SYBR Premix Ex Taq II (10 μL), forward and reverse primer (0.5μL), Rox Reference Dye (1μL), cDNA template (2μL), and distilled water (6μL) was prepared in the final volume of 20 μL. The U6 forward and reverse primers were also provided by the commercial kit. We used the Rotorgene RealTime PCR system for the qRT-PCR reactions and performed the tests in duplicate (Table 1). U6 (RNU6‑1) snRNA were used as reference genes for patients and control samples. The efficiency for mir-222 real-time PCR was 0.95, and R^2 was 0.991. The efficiency for the reference U6 real-time PCR was 0.96, and R^2 was 0.982
Statistical Analysis
For data analysis, we used SPSS software version 22 and GraphPad Prism version 8.4.3 (Graph Pad Software Inc., San Diego, CA, USA). The comparisons between groups were analyzed using Mann-Whitney 𝑈 test, Pearson X2 test, or Spearman correlation analysis where appropriate. A p-value less than 0.05 was considered statistically significant.
Table 1. qRT-PCR protocol for relative quantification of miR-222
| 1 cycle | 45 cycle | 1 cycle | |||
| Melt curve | Annealing and Extension | Denaturation | Primary denaturation | ||
| 95℃ | 60℃ | 95℃ | 60℃ | 95℃ | 95℃ |
| 15 Sec | 10 sec | 15 sec | 34 sec | 5 sec | 30 sec |
The expression level of miR-222 in the patient and control groups was determined using the qRT-PCR method and analyzed the results with SPSS software. Individuals with a mean age of 38.09±13.52 for the patient group and 36.63±12.23 for the control group of both males and females were studied, and alanine transaminase levels (ALT) and aspartate transaminase (AST) enzymes were measured. Table 2 summarizes the demographic parameters of the patient and control group.
In the case of miR-222 (miRNA-222 Fold change= 1.384, p value=0.269) despite the difference in expression levels between the patient and control groups, we did not observe a significant difference (figure1).
Table 2. Demographic and clinical parameters of the study participants
| Characteristics | Patients (Mean ± SD) | Healthy controls (Mean ± SD) | p-value |
| Age (years) | 38.09±13.52 | 36.63±12.23 | 0.539 |
| Gender/Male | 23(53.49%) | 24(55.81%) | 0.829 |
| Gender/female | 20(46.51%) | 19(44.19%) | |
| ALT (U/L) | 43.49+50.18 | 21.28+9.05 | <0.001 |
| AST (U/L) | 31.88+27.98 | 18.00+6.01 | 0.036 |
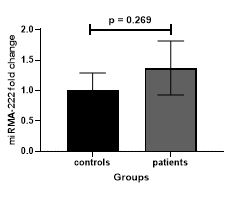
Figure 1. Comparison of miR-222 expression between patient and control groups (miRNA-222 Fold change= 1.384, p value=0.269).
The liver is the main organ performing many vital bodily functions (32). Studies have shown that miRNAs can affect liver function, and thus changes in these molecules can affect the functional network of the liver and lead to liver disease (33). MiRNAs, such as the other molecules, including kinase inhibitors, antibodies, and siRNAs, have been used to suppress the function of crucial signaling pathways (34, 35). So far, several studies have been published on the association of miRNAs expression with various liver diseases, including chronic hepatitis B. For example, the relationship between the expression level of miR-29a, miR-15a/miR-16-1, let-7a, let-7b, let-7c, miR-122, miR-572, miR-575, miR -638, miR-885-5p, miR145, miR-21, miR-222, miRNA-146a, miR-744, miR-101 with chronic hepatitis B has been studied (36-45). MiR-222, one of the miRNAs studied in liver diseases, controls the activation of Akt by regulating the expression of PTEN, a tumor suppressor gene that acts upstream of Akt. It has been mentioned in some studies that Mir-222 is involved in the promotion of an aggressive basal-like phenotype in some types of cancers through functioning downstream of the RAS pathway (46).
Motawi et al., studied patients with liver cancer and chronic hepatitis C and found that serum levels of miR-222 in hepatitis C patients should increase significantly, and this miRNA can be considered a diagnostic biomarker for liver damage (47). In a study, peng et al., checked out mir-222 as a biomarker for hepatocellular Carcinoma in Chinese Patients with Chronic Hepatitis B Virus Infection and found that it was significantly up-regulated between healthy controls and HCC patients. However, no significant difference was observed in the levels of mir-222 between HBV patients without and with HCC (48). A study by cheng et al., investigated the potential of serum miRNAs like miR-222 as novel biomarkers for hepatocellular carcinoma. They demonstrated that miR-222 was significantly change regulated between HCC patients and the control group, and no significant difference was observed in the levels of miR-222 between HBV patients with and without HCC (49).
Ladeiro et al., investigated the expression levels of miR-222 in benign and malignant hepatocellular tumors and showed the miR-222 deregulation in these cases (50).
Some studies shown increasing expression and up-regulation of miR-222 in patients with human hepatocellular cancer (50-52). In a study, Bandopadhyay et al., investigated the low expression of miR-222 in HepG2 cells transfected with HBX protein (53).
In a study, Sasaki et al., were analyzed the expression of miR-222 in the liver and serum of patients with fibrosis and cirrhosis and found that this microRNA caused the proliferation and differentiation of hepatic stellate cells (HSCs) in the liver and its expression in both advanced fibrosis and cirrhosis was increased compared to mild fibrosis and cirrhosis. Furthermore, miR-222 downregulates the expression of ICAM-1 28, which transports immune cells to the site of liver damage. The up-regulation of miR-222 led to an acceleration of HSCs activation, increased the liver's inflammation, and eventually facilitated the progression of fibrosis (54).
Cheng et al., in a study, demonstrated the effect of hepatitis E virus regulatory protein (Swine HEV) on cytoplasmic signaling pathways. They showed that the cyclin-dependent kinase inhibitor (p27) is regulated directly by the miR-222, and miR-222 downregulation caused by the hepatitis E virus leads to increased expression of the target gene in HEK293 cells (55). Ogawa et al demonstrated miR-222 expression in hepatitis patients and the inhibitory effect of NF-κB. They showed that the inhibitor significantly suppressed miR-222 induction. They also showed that the miR-222 could be considered a marker in the progression of liver fibrosis (56).
In a study by Yang et al., on the abnormal expression of miRNA molecules in patients with chronic hepatitis B, they found that molecular pathways and target genes regulate immune clearance pathways in patients with chronic hepatitis B. They also reviewed 221 miRNAs, including miR-222, in these patients and reported reduced levels of miR-222 expression in patients with chronic hepatitis B (57).
In a study, Sohn et al., demonstrated that the serum levels of miR-222 were significantly higher in patients with HCC than those with CHB (58). Upregulation of miR-222 was observed in primary HCC compared with normal liver tissue in a study by Wong et al. (59).
Lendvai et al., measured the level of miR-222 expression in patients with focal nodular hyperplasia (FNH), cirrhosis and HCC compared to healthy liver tissue and showed that the level of miR-222 expression, unlike FNH, was high in cirrhosis and HCC. They concluded that this could be related to the malignancy process (60).
Yang et al shown that overexpression of miR-222 increased the proliferation of HepG2 cells by downregulation of the p27 gene, thereby explaining the mechanism of the association of miR-222 with HCC (61). Zhang et al., showed a high level of mir-222 in HCC patients compared to the control group (62).
Simao et al., demonstrated the function of mitochondria-targeting miRNAs, including miR-222, in metabolic syndrome-associated NAFLD and shown liver miR-222-3p-induced mitochondrial dysfunction is significant as a therapeutic target (63).
Ying et al., in a study on patients with CHB-related liver fibrosis, showed that the level of miR-222 expression in these patients had increased significantly and, with further studies, can be a notable candidate as a diagnostic biomarker (64). We also examined the expression of miR-222 in patients with chronic hepatitis B. We concluded that despite the increase in the expression of this miRNA in the patient group (Fold change: 1.384), no significant difference was observed.
In this study, we demonstrated an increase in the expression level of miR-222 in patients with chronic hepatitis B compared to the control group. But despite that, the difference was not statistically significant. Although miR-222 is one of the miRNAs involved in influencing the pathways leading to liver activity, limited studies on its role in CHB as a noninvasive biomarker have been performed. Because this miRNA is effective in the proliferation of liver cells and on the part of molecular pathways, it reduces the presence of immune cells at the site of liver damage, it can play a role in liver pathogenesis, and its expression change is considered as a biomarker. Further studies on miR-222 are needed to determine the potential role of this miRNA and its ability as a diagnostic marker in patients with chronic hepatitis B.
This research was funded by the Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran. The authors thank all Gastroenterology and Liver Diseases Research Center staff for their enthusiastic efforts, especially Mrs. Shabnam Kazemian, Mrs. Mahsa Saeedi, and Mrs. Farahnaz Jabarian.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Received: 2022/05/19 | Accepted: 2022/07/3 | ePublished: 2022/09/9
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |





