BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1745-en.html


 , Ramin Rezaee2
, Ramin Rezaee2 

 , Mohammad Hashemi1
, Mohammad Hashemi1 

 , Behzad Kiani3
, Behzad Kiani3 

 , Sajjad Ghasemi4
, Sajjad Ghasemi4 

 , Mahmood Alizadeh Sani5
, Mahmood Alizadeh Sani5 

 , Asma Afshari6
, Asma Afshari6 


2- UNESCO International Center for Basic Sciences Related to Human Health and Nutrition, Faculty of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran
3- Department of Medical Informatics, Faculty of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran
4- Department of Food Science and Technology, Faculty of Agriculture, Ferdowsi University of Mashhad, Mashhad, Iran
5- Division of Food Safety and Hygiene, Department of Environmental Health Engineering, School of Public Health, Tehran University of Medical Sciences, Tehran, Iran
6- Department of Nutrition, Faculty of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran ,
Staphylococcal food poisoning has been introduced as the third cause of food-borne disease throughout the world (1). Staphylococcus aureus is one of the most worrying pathogens in food due to the ability of some strains to produce heat-stable enterotoxins that can produce gastrointestinal disorders (2). Also, as an opportunistic pathogen for both humans and animals, it can cause pneumonia, osteomyelitis, brain abscesses, toxic shock syndrome, wound infections, bacteremia, and meningitis (3).
Enterococcus bacteria are naturally present in the gastrointestinal microflora of warm-blooded animals and humans (4). For many years, Enterococcus spp. were believed to be harmless to humans and medically unimportant (5). However, these bacteria can cause hospital-acquired infections such as urinary tract and wound infections, bacteremia, and endocarditis (4).
Besides toxic effect of foodborne bacterial pathogens, their resistance to antibiotics remains a major threat for animal and human health and its prevalence has increased in recent decades (6). The World Health Organization (WHO) introduced the issue of antimicrobial resistance as one of the biggest health challenges of the 21th century (7). Foodstuffs are an important vehicle for transmission of antibiotic-resistant bacterial strains to the gastrointestinal tract of consumers (8-10). Livestock and livestock-derived foods can serve as a reservoir for human infections caused by antibiotic-resistant Staphylococcus strains (1) and potentially transfer resistant Enterococcus species (11). In fact, the over-use and often irrational use of prophylactic antibiotics accelerate the emergence and spread of drug-resistant strains (12).
A well-known player in the antimicrobial resistance crisis is methicillin-resistant S. aureus (MRSA) (13). Multi-drug resistance in pathogenic bacteria was first mentioned in the 1950s (14). After emergence and dissemination of MRSA strains, vancomycin, a glycopeptide antibiotic, was used for the treatment of infections caused by methicillin-resistant Staphylococci. In 1996, vancomycin-resistant S. aureus strains (VRSA) emerged in Japan. Because vancomycin is not regularly used for treating infections in animals, few reports on VRSA strains in veterinary medicine exist (15). Vancomycin-resistant Enterococcus (VRE) (more frequently seen for E. faecium and less for E. faecalis) has a lower epidemiological impact compared to MRSA and they are almost exclusively restricted to healthcare settings (13).
Methicillin-Resistant Staphylococcus aureus (MRSA)
From different parts of the world, MRSA species have shown resistance towards first-line antibiotics including beta-lactams (16). MRSA is known to be resistant against almost all types of penicillin and β-lactam antibiotics (17). Methicillin resistance in staphylococci is probably the most mysterious among all bacterial antibiotic resistance (18). Mechanism of the intrinsic form of penicillin resistance in S. aureus is unknown (18, 19). Methicillin resistance in S. aureus has been considered the most important clinical resistance trait (20).
During the past years, the rate of occurrence of infections caused by MRSA significantly increased in livestock (i.e., livestock-associated MRSA (LA-MRSA)) (3). LA-MRSA may also be transmitted to humans through occupational contact with infected livestock and consumption of foods prepared from contaminated animals (3, 21). Animals are recognized as the main source of new pathogenic strains of S. aureus for human, particularly the clones which possess no host specificity (3). The probability of MRSA transmission via food was largely unknown until 1994 (22). MRS isolates were initially reported in dairy animals in 1972 (from mastitis cows) (16).
Vancomycin-Resistant S. aureus (VRSA)
Following the advent of MRSA strains and their rapidly increasing resistance to different groups of antibiotics (like penicillin, cephalosporins, aminoglycosides, fluoroquinolones, and macrolides), vancomycin was used for treatment of patients infected with MRSA or other gram-positive bacteria (23). The first strains of S. aureus with reduced sensitivity to vancomycin were reported in Japan in 1997 (24). Thereafter, VRSA as well as vancomycin intermediate-resistant S. aureus (VISA) were identified in the United States, the United Kingdom, Germany, Portugal, Brazil, China, Bangladesh, and Jordan (23).
Only a few reports were published about vancomycin-resistant strains of S. aureus in veterinary medicine because vancomycin is not regularly used in the treatment of infected animals. The emergence of such strains in animals could be due to the contamination of pasture soils or environment with vancomycin-resistant isolates originating from patients who had been to a healthcare facility and resided very close to animals and milching of milch animal (15).
Vancomycin-Resistant Enterococci (VRE)
Vancomycin-resistant enterococci (VRE) were first recognized in the United Kingdom and France in 1986 (25, 26). The emergence of VRE was reported to be related to severe clinical use in the US and cross resistance in Europe, and subsequently, the agricultural use of avoparcin as a growth promoter (25).
In developing countries, antibiotics and anthelmintics (compounds such as antibiotics, coccidiostats, and growth promoting hormones) are widely used to control the pathogens and satisfy the increased demand for meat and dairy products. Hence, some studies in Iran focused on the prevalence of MRSA, VRSA, and VRE in foodstuffs. The objective of this review was to collect and thoroughly review the available information regarding the prevalence of MRSA, VRSA, and VRE in different food samples of animal and plant origin in Iran.
Search Strategy
PubMed, ScienceDirect, Scopus, SID, and Google Scholar were searched for articles published between 2006 and September 2020 using the following keywords: “Staphylococcus aureus”, “Food”, “Enterococcus”, “Antimicrobial resistance”, “Prevalence”, “Iran”, “MRSA”, “VRSA”, and “VRE”. Initial screening of the retrieved articles was done through reading their titles and/or abstracts. Sixty-five papers were selected, of which 23 articles including duplicate publications, conference abstracts, and studies that did not contain complete information were excluded. The location of the studies was geocoded using the Google My Maps (https://www.google.com/mymaps) software. The ArcMap 10.6 software was used to manage the geographic data, create maps, and provide an overview of the geographical distribution of MRSA contamination in six different food categories including milk, cheese, other dairy products, pastry products, meat products, and miscellaneous foods and VRE in miscellaneous foods in different regions of Iran.
Meta-Analysis
The overall prevalence meta-analysis was calculated by using “meta prop program” in STATA statistical software (STATA, College Station, TX, USA, version 15.0). To estimate the pooled prevalence and corresponding 95% confidence interval (CI), the random-effects model was applied. Cochran's Q test and I2 were used to determine statistical heterogeneity between studies. The meta regression test was used to evaluate possible sources of heterogeneity based on the duration and location of the study. P < 0.05 was considered as a statistically significant.
Presence of MRSA in Various Food Samples
The number of articles about the prevalence of MRSA strains in Iran has increased in recent years. However, the available information is limited to reports from a few cities. Most of these studies are focused on clinical isolates but there are some papers in which strains from various food samples were analyzed. MRSA strains were identified in milk, cheese, butter, yoghurt, ice cream, Kashk, pastry, meat, fish, and ready-to-eat food samples in some Iranian studies (Tables 1-6 and Figures 1-6). Several studies done in different regions of Iran examined the presence of MRSA in milk (Table 1). Many of these MRSA strains were identified using molecular typing techniques. The molecular identification (using polymerase chain reaction (PCR)) of MRSA strains was performed by investigating mecA gene. In these studies, antibiotic resistance pattern of isolated MRSA strains was also tested (27-32). In addition, some studies found MRSA strains in milk by phenotypic methods (disk diffusion method) (33, 34).
Based on the literature, so far, MRSA strains were reported in milk samples in 6 provinces and in 4 of them (Chaharmahal and Bakhtiari, East Azerbaijan, Hamadan, and Mazandaran) MRSA strains were found. The MRSA prevalence identified in the different provinces was highly variable (from 0.00% to 13.17%; Figure 1). Chaharmahal and Bakhtiari province showed the highest prevalence (11.51%-13.7%). Nonetheless, MRSA strains were not identified in milk samples from Fars province.

Figure 1. Distribution of methicillin-resistant Staphylococcus aureus (MRSA) prevalence in milk samples in Iran during 2006-2020.
Table 1. Reports on the presence of methicillin-resistant Staphylococcus aureus (MRSA) in milk samples in Iran during 2006-2020.
| Milk | ||||||
| Sample type | Location | No. of samples | No. of S. aureus positive samples (Prevalence) |
No. of MRSA positive samples (Prevalence) |
Year | Reference |
| Raw cow milk | Chaharmahal and Bakhtiari (Shahr-e Kord) | 152 | 89 (58.55%) | 68 (44.73%) | 2016 | (27) |
| Raw sheep milk | Chaharmahal and Bakhtiari (Shahr-e Kord) | 32 | 18 (56.25%) | 10 (31.25%) | 2016 | (27) |
| Raw goat milk | Chaharmahal and Bakhtiari (Shahr-e Kord) | 52 | 16 (30.76%) | 11 (21.15%) | 2016 | (27) |
| Pasteurized cow milk | Chaharmahal and Bakhtiari (Shahr-e Kord) | 65 | ND | ND | 2016 | (27) |
| Raw milk | Hamadan | 131 | NS | 2 (1.52%) | 2013-2014 | (28) |
| Pasteurized milk | East Azerbaijan (Tabriz) | 100 | 1 (1%) | ND | 2010 | (32) |
| Raw milk | East Azerbaijan (Tabriz) | 100 | 45 (45%) | 14 (14%) | 2010 | (32) |
| Raw cow milk | Mazandaran | 1035 | 162 (15.7%) | 21 (2.02%) | 2006-2013 | (29) |
| Raw sheep milk | Mazandaran | 895 | 86 (9.6%) | 11 (1.23%) | 2006-2013 | (29) |
| Raw milk | Mazandaran | 120 | NS | 44 (36.67%) | 2017-2018 | (30) |
| Raw milk | Hamadan | 271 | 29 (10.7%) | 3 (1.107%) | 2013-2014 | (31) |
| Raw milk (cow, sheep and goat) | Fars, Chahar Mahal va Bakhtiari and Ghom | 348 | 46 (13.2%) | ND | 2010-2011 | (33) |
| Raw milk | NS | 100 | 60 (60%) | 31 (31%) | NS | (34) |
ND = Not Detected (it means that the referred bacteria was not detected in samples); NS = Not Specified (it means that the referred item was not specified in the article by the author).
Numerous studies evaluated the prevalence of MRSA in cheese samples in different provinces of Iran (Table 2). In most studies, MRSA detection was done using specific mecA primers, by PCR method (27, 28, 31, 35-40). The MRSA prevalence was identified in different provinces including Mazandaran, Hamadan, Khorasan Razavi, East Azerbaijan, West Azerbaijan, and Chaharmahal and Bakhtiari (Table 2 and Figure 2). A high variation in MRSA prevalence in cheese samples among different provinces and even in cheese samples from the same province was observed (Table 2). As shown in Figure 2, the prevalence of MRSA in cheese samples varied from 0.01 to 22.22 with Chaharmahal and Bakhtiari province having the highest prevalence (17.26% -22.22%) and Hamadan having the lowest prevalence (0.01-5.75).
Differences in results of different studies can be attributed to variations in cheese production technologies, number of samples, milk source (raw/pasteurized), and hygiene measures which impact the rate of contamination.
Table 2. Reports on the presence of methicillin-resistant Staphylococcus aureus (MRSA) in cheese in Iran during 2006-2020.
| Cheese | ||||||
| Sample type | Location | No. of samples | No. of S. aureus positive samples (Prevalence) |
No. of MRSA positive samples (Prevalence) |
Year | Reference |
| Traditional cheese | Mazandaran | 360 | 224 (62.2%) | 119 (33.05%) | 2016-2017 | (36) |
| Traditional cheese | Mazandaran | 450 | 49 (10.9%) | 15 (3.33%) | 2006-2013 | (29) |
| Traditional and industrial white cheese | Hamadan | 120 | 19 (15.8%) | 4 (3.33%) | 2015 | (64) |
| Cheeses (white and feta) | Khorasan Razavi (Mashhad) | 100 | 25 (25%) | 8 (8%) | NS | (35) |
| Traditional cheeses | East Azerbaijan (Tabriz) | 100 | 19 (19%) | 9 (9%) | NS | (38) |
| Traditional cheese | West Azerbaijan (Qotur of khoy) | 80 | 43 (53.57%) | 4 (5%) | 2011 | (65) |
| Traditional cheese |
Chaharmahal and Bakhtiari (Shahr-e Kord) | 27 | 11 (40.74%) | 6 (22.22%) | 2016 | (27) |
| Traditional cheeses | Azerbaijan | 100 | 16 (16%) | 3.36 (3.36%) | 2012 | (39) |
| Traditional cheese | West Azarbaijan and East Azarbaijan | 73 | 123** (NS) | 19** (15.44%) | NS | (40) |
| Local cheese | East Azerbaijan | 100 | 22 (22%) | 1 (1%) | 2017 | (37) |
| Cheese | Hamadan | 170 | 19 (11.2%) | 2 (1.17%) | 2013-2014 | (31) |
| Cheese | Hamadan | 84 | NS | 1 (1.19%) | 2013-2014 | (28) |
NS = Not Specified (it means that the referred item was not specified in the article by the author); ** Numbers of isolates.
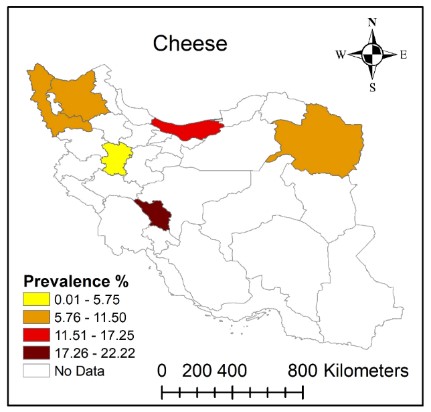
Figure 2. Distribution of methicillin-resistant Staphylococcus aureus (MRSA) prevalence in cheese samples in Iran during 2006-2020.
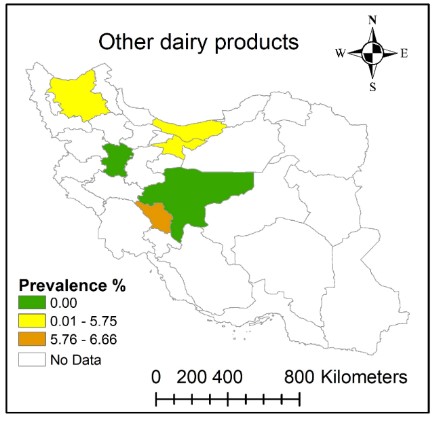
Several studies surveyed the prevalence of MRSA in milk and cheese while few studies considered other dairy products (Table 3). MRSA identification was done through mecA gene detection (27, 29, 31, 32, 38, 41, 42). Different dairy products including butter, yoghurt, cream/ top of the milk, ice cream, and Kashk were evaluated in Chaharmahal and Bakhtiari, Mazandaran, Hamadan, East Azerbaijan, Tehran, and Isfahan (Table 3). The prevalence of MRSA varied between 0.00 and 6.66 (Figure 3). A high prevalence of MRSA was reported in Chaharmahal and Bakhtiari while MRSA strains were not detected in different dairy product samples from Hamadan (Figure 3). MRSA strains were identified in ice cream samples in Tehran but were not found in ice cream samples from Chaharmahal and Bakhtiari, Hamadan, and Isfahan. Different sampling methods, seasonal effects, and laboratory techniques could have caused the observed differences in the results among different studies.
Table 3. Reports on the presence of methicillin-resistant Staphylococcus aureus (MRSA) in other dairy products in Iran during 2006-2020.
| Other Dairy Products | ||||||
| Sample type | Location | No. of samples | No. of S. aureus positive samples (Prevalence) |
No. of MRSA positive samples (Prevalence) |
Year | Reference |
| Traditional butter | Chaharmahal and Bakhtiari (Shahr-e Kord) | 30 | 7 (23.33%) | 3 (10%) | 2016 | (27) |
| Traditional yoghurt | Chaharmahal and Bakhtiari (Shahr-e Kord) |
15 | 2 (13.33) | 1 (6.66%) | 2016 | (27) |
| Traditional ice cream | Chaharmahal and Bakhtiari (Shahr-e Kord) |
18 | 6 (33.33%) | ND | 2016 | (27) |
| Traditional “Kashk” ** | Chaharmahal and Bakhtiari (Shahr-e Kord) |
12 | 2 (16.66%) | 1 (8.33%) | 2016 | (27) |
| “Kashk” | Mazandaran | 270 | 31 (11.5%) | 6 (2.22%) | 2006-2013 | (29) |
| Cream | Hamadan | 66 | 4 (6.1%) | ND | 2013-2014 | (31) |
| Traditional yoghurt | Hamadan | 45 | 2 (4.4%) | ND | 2013-2014 | (31) |
| Traditional cream | Hamadan | 47 | 2 (4.3%) | ND | 2013-2014 | (31) |
| Butter | Hamadan | 72 | 16 (8.3%) | ND | 2013-2014 | (31) |
| Traditional butter | East Azerbaijan (Tabriz) | 150 | 11 (7.33%) | 2 (1.33%) | NS | (38) |
| Ice cream | East Azerbaijan (Tabriz) | 100 | 23 (23%) | 6 (6%) | 2010 | (32) |
| Cream | Hamadan | 32 | NS | ND | 2013-2014 | (28) |
| Yoghurt | Hamadan | 21 | NS | ND | 2013-2014 | (28) |
| Top of the milk | Hamadan | 22 | NS | ND | 2013-2014 | (28) |
| Butter | Hamadan | 35 | NS | ND | 2013-2014 | (28) |
| Ice cream | Tehran | 241 | 41 (17%) | 2 (0.83%) | 2006-2007 | (66) |
| Traditional and commercial dairy products |
Isfahan | 347 | 20 (5.8%) | ND | 2010-2011 | (67) |
ND = Not Detected (it means that the referred bacteria was not detected in samples); NS = Not Specified (it means that the referred item was not specified in the article by the author); ** “Kashk” is fermented and dried cow’s and/or sheep’s milk used in Iranian diets with a salty or sour taste.
Studies on the isolation of MRSA from pastry products are shown in Table 4. These studies reported MRSA in pastry cream in Mazandaran and Tehran provinces and in sweet samples in Hamadan province (Table 4) as examined by PCR to detect mecA (Azizkhani and Tooryan 2018) or nuc genes (Fatahi et al. 2018) and disc diffusion method (Soltan Dallal et al. 2008). Prevalence of MRSA in pastry products varied among different regions (0.01%-9.46%; Figure 4). The highest prevalence of MRSA in pastry products was reported in Hamadan followed by Mazandaran and Tehran provinces (Table 4 and Figure 4). According to a study conducted in Mazandaran (Amol), remarkable differences in the prevalence of MRSA may be due to the providing optimum temperature for S. aureus growth in warm seasons (36).

Figure 4. Distribution of methicillin-resistant Staphylococcus aureus (MRSA) prevalence in pastry products in Iran during 2006-2020.
Table 4. Reports on the presence of methicillin-resistant Staphylococcus aureus (MRSA) in pastry products in Iran during 2006-2020.
| Pastry Products | ||||||
| Sample type | Location | No. of samples | No. of S. aureus positive samples (Prevalence) |
No. of MRSA positive samples (Prevalence) |
Year | Reference |
| Pastry cream | Mazandaran (Amol) | 360 | 150 (41.6%) | 11 (3.05%) | 2016-2017 | (36) |
| Sweet | Hamadan | 370 | 100 (27.02%) | 35 (9.46%) | 2017-2018 | (68) |
| Pastry cream | Tehran | 214 | 37 (17.3%) | 1 (0.46%) | 2006-2007 | (66) |
NS = Not Specified (it means that the referred item was not specified in the article by the author).
Studies that examined meat and meat products for MRSA contamination are shown in Table 5. In these studies, molecular detection of specific target of MRSA, mecA gene, was performed for all S. aureus strains isolated from meat and meat products in Tehran, Isfahan, and Hamadan provinces (28, 31, 41-49). The prevalence of MRSA in meat and meat products varied from 0.01% to 19.44% (Figure 5). Isfahan had the highest prevalence of MRSA (17.26%-19.44%). Prevalence of MRSA in these products varied between 0.01% and 5.75%, while MRSA strains were not identified in meat samples from Hamadan province (Figure 5). The upper line differences observed among studies may be related to the type of meat processing, human hygiene-related factors, sample preparation, and analysis techniques.

Figure 5. Distribution of methicillin-resistant Staphylococcus aureus (MRSA) prevalence in meat and meat products in Iran during 2006-2020.
Table 5. Reports on the presence of methicillin-resistant Staphylococcus aureus (MRSA) in meat and meat products in Iran during 2006-2020.
| Meats | ||||||
| Sample type | Location | No. of samples | No. of S. aureus positive samples (Prevalence) |
No. of MRSA positive samples (Prevalence) |
Year | Reference |
| Packaged hamburgers | Tehran | 256 | 64 (25%) | 58 (22.65%) | 2010 | (48) |
| Raw meat (beef, chicken, turkey) | Tehran | 131 | NS | 49 (37.4%) | 2016 | (47) |
| Chicken meat | Isfahan | 36 | NS | 25 (69.4%) | 2014 | (42) |
| Raw chicken meat | Isfahan | 360 | 82 (22.77%) | 68 (18.8%) | 2011-2012 | (46) |
| Raw meat (beef, sheep, goat, and camel) | Isfahan | 900 | NS | 160 (17.7%) | 2011-2012 | (46) |
| Raw red meat | Hamadan | 243 | 25 (10.3%) | ND | 2013-2014 | (31) |
| Raw poultry | Hamadan | 136 | 11 (8.1%) | ND | 2013-2014 | (31) |
| Red meat | Hamadan | 119 | NS | ND | 2013-2014 | (28) |
| Chicken meat | Hamadan | 66 | NS | ND | 2013-2014 | (28) |
| Protein food (raw and cooked) | Tehran | 481 | 18 (3.7%) | ND | 2006-2007 | (66) |
| Shrimp (fresh and frozen) | Tehran | 300 | 84 (28%) | 24 (8%) | 2013-2014 | (49) |
| Fish and shrimp (fresh and frozen, marine and farmed) | Tehran | 600 | 206 (34.3%) | 49 (8.16%) | 2013-2014 | (43) |
ND = Not Detected (it means that the referred bacteria was not detected in samples); NS = Not Specified (it means that the referred item was not specified in the article by the author).
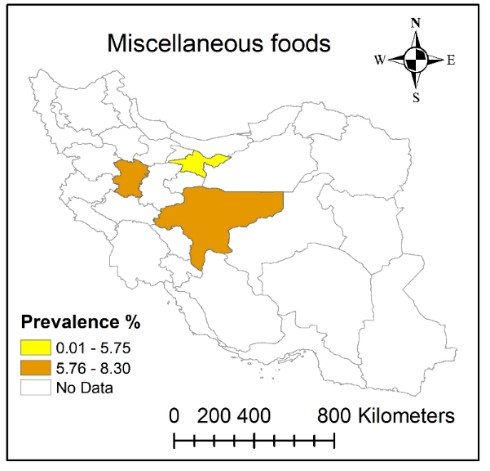
Studies that examined MRSA in miscellaneous foods are listed in Table 6. Some of these studies tested for the presence of MRSA strains using PCR to detect mecA gene (41, 50-54). In a study done in Isfahan, a high prevalence of MRSA strains was found in the restaurant food samples. Cooked chicken samples had the highest prevalence of MRSA strains. No study reported MRSA strains in raw fish. Low prevalence of MRSA in fish meat samples was attributed to the low ability of S. aureus to compete with specific primary bacterial flora of fish (53). In another study, MRSA was most prevalent in meat barbecue, chicken barbecue, soup, and salad followed by raw chicken meat, raw read meat, and rice. Also, MRSA strains were not found in raw fish and grilled fish (50). According to Table 6, the prevalence of MRSA in provinces of Isfahan, Tehran, and Hamadan was within the range of 0.01%-8.30% (Figure 6). The highest prevalence of MRSA was identified in restaurant food in Isfahan province (55) followed by Samosa and Falafel in Hamadan province (56) and hospital food in Isfahan (57) (Table 6). So, it seems that foods prepared in public places including restaurants, hospitals, and sandwich shops are among the most common sources of foodborne MRSA. As shown in Figure 6, the prevalence of MRSA in samples from Isfahan and Hamadan provinces was higher (5.76%-8.30%) compared to Tehran province (0.01%-5.75%).
Figure 6. Distribution of methicillin-resistant Staphylococcus aureus (MRSA) prevalence in miscellaneous foods in Iran during 2006-2020.
Table 6. Reports on the presence of methicillin-resistant Staphylococcus aureus (MRSA) in miscellaneous foods in Iran during 2006-2020.
| Miscellaneous Foods | ||||||
| Sample type | Location | No. of samples | No. of S. aureus positive samples (Prevalence) |
No. of MRSA positive samples (Prevalence) |
Year | Reference |
| Ready to eat foods | Isfahan | 384 | 4 (1.042%) | 4 (1.04%) | 2015 | (59) |
| Restaurant food | Isfahan | 580 | 119 (20.51%) | 83 (14.31%) | 2015-2016 | (53) |
| Dairy and meat products | Isfahan | 139 | 9 (6.48%) | 0.86 (0.61%) | 2015-2016 | (51) |
| Hospital food | Isfahan | 485 | 47 (9.69%) | 37 (7.62%) | 2015-2016 | (50) |
| Hospital food | Tehran | 44 | 2 (4.54%) | 1 (2.27%) | NS | (69) |
| Fruit juice | Tehran | 32 | 3 (9.4%) | ND | 2006-2007 | (66) |
| Salad | Tehran | 79 | 1 (1.3%) | ND | 2006-2007 | (66) |
| Dairy and meat products | Tehran | 913 | 93 (10.18%) | 5 (0.54%) | 2010 | (54) |
| Samosa and Falafel | Hamadan | 120 | 56 (46.67%) | 10 (8.3%) | 2015-2016 | (52) |
ND = Not Detected (it means that the referred bacteria was not detected in samples); NS = Not Specified (it means that the referred item was not specified in the article by the author).
Presence of VRSA in Various Food Samples
Only one study in Iran has investigated the prevalence of VRSA strains in food samples. Totally, 119 samples of chicken and turkey raw meat were analyzed for the presence of Staphylococcus aureus. Based on this report, 29 strains of S. aureus were isolated; out of them, 14 (48.5%) strains were found to be resistant to vancomycin as shown by phenotypic methods. The presence of vanA gene using molecular typing techniques (PCR) was identified in chicken and turkey raw meat samples. The prevalence of vanA gene in the tested food samples was 43% (6 strains) (58). In some studies, performed in Iran, vancomycin-resistant strains were identified by phenotypic methods but the presence of van gene was not studied (32, 36, 38, 48, 53, 59).
Presence of VRE in Various Food Samples
Iranian studies that assessed the prevalence of VRE in different food samples are presented in Table 7. In a study conducted in Tehran province, vancomycin-resistance was most-commonly observed for E. faecalis isolates followed by E. faecium, E. gallinarum, and E. durans. The prevalence of five of the van (vanA, B, C, D, and E) genes was investigated by PCR technique. VanA, vanB and vanC genes were the most common phenotypes observed in the isolates, respectively, while vanD and vanE genes were not identified. The findings of this study suggest that the presence of Enterococci in packed and unpacked dried vegetables is due to application of contaminated organic fertilizers and irrigation with wastewater (45). In another study in Tehran, PCR-based assay showed that all of the VRE isolates were E. faecium. All of the isolates were positive for vanA, but vanB was not detected. Isolates displaying positive results for VRE were observed in meat, chicken, and cheese samples, respectively. The results of this study indicated a high prevalence of VRE in meat and chicken possibly due to extensive use of glycopeptide antibiotics including vancomycin in veterinary practices (60). In a study performed in Hamadan, VRE strains were found in chicken meat, raw ground beef (14 and 27 strains), milk, and cheese (6 and 1 strains). VanA, vanB, and vanC were detected in VRE isolates (61). In a study conducted in Lorestan province, vanC type was not identified by PCR (62). The highest prevalence of VRE was found in Ilam province, E. faecalis was recovered from all food samples. Also, all E. faecalis isolates showed resistance to vancomycin (63). As shown in Figure 7, variable prevalence of VRE (0.00% to 100.0%) was reported from different provinces of Iran. Ilam and Lorestan provinces had the highest prevalence of VRE (75.01-100.00). In Tehran, VRE prevalence was 25.01-50.00 and in Hamadan province it was 0.001-25.00. No VRE strain was detected in dairy product samples from Urmia and Tabriz (East Azerbaijan and West Azerbaijan provinces, respectively).

Figure 7. Distribution of vancomycin-resistant enterococci (VRE) prevalence in miscellaneous food samples in Iran during 2006-2020.
Table 7. Reports on the presence of vancomycin-resistant enterococci (VRE) in miscellaneous food samples in Iran during 2008-2020.
| Miscellaneous Food Samples | ||||||
| Sample type | Location | No. of samples | No. of Enterococcus positive samples (Prevalence) |
No. of VRE positive samples (Prevalence) |
Year | Reference |
| Packed and unpacked dried vegetables | Tehran | 140 | 84 (60%) | 41 (29.28%) | 2015 | (45) |
| Meat, chicken, cheese | Tehran | 30 | NS | NS | 2010 | (60) |
| Dairy products and meat | Hamadan | 200 | 135 (67.5%) | 48 (24%) | 2012-2014 | (61) |
| Ground meat, raw meat, kebab | Ilam | 34 | 34 (NS) | 34 (NS) | NS | (63) |
| Red meat | Lorestan (Borujerd) | 181** | 181** | 68** (83.95%) | 2014-2015 | (62) |
| Traditional cheese | Urmia and Tabriz | 50 | 48 (96%) | ND | 2014-2015 | (70) |
NS = Not Specified (it means that the referred item was not specified in the article by the author); ** Numbers of isolates.
Meta-Analysis Results
Forest plot diagram of the current systematic review and meta-analysis based on overall prevalence of MRSA, VRSA, and VRE in foods are shown in Supplementary file. In addition, meta-regression plot of the current systematic review and meta-analysis in foods based on year of studies were indicated in Supplementary file. Funnel plot and Sensitivity of the current systematic review and meta-analysis in foods were also displayed in Supplementary file. Moreover, the pooled ES of MRSA in milk, cheese, other dairy product, pastry product, meat product, and miscellaneous foods were as follows, respectively: 0.26% (95% CI: 0.19%, 0.34%), 0.25% (95% CI: 0.13%, 0.36%), 0.12% (95% CI: 0.08%, 0.16%), 0.29% (95% CI: 0.15%, 0.42%), 0.21% (95% CI: 0.13%, 0.29%), and 0.11% (95% CI: 0.06%, 0.17%) (Supplementary file). Accordingly, the highest pooled ES of MRSA was for pastry products and the lowest pooled ES of MRSA was for miscellaneous foods. Furthermore, the pooled ES of VRE in miscellaneous foods was 0.80% (95% CI: 0.62%, 0.98%) (Supplementary file).
Antimicrobial resistance is one of the most important threats to the public health and food safety and security. Since food plays a critical role in development of antimicrobial resistance, identification of food sources and geographical distribution patterns is necessary to control and manage antimicrobial resistance. This review demonstrated that antimicrobial resistance varies among different foodstuffs. Such disparities can be attributed to differences in food handling practices, hygiene practices during processing, traditional food-processing and preparation methods, geographical area and use of antimicrobials in plant and animal agriculture. Based on the published reports, MRSA, VRSA and VRE strains are widely present in different types of raw and processed meat, raw milk, traditional cheese, restaurant, and hospital foods. Of note, preventing excessive use of antimicrobials can be helpful in guaranteeing the product safety from farm to fork. Good agricultural practices, preventive controls, surveillance system in agriculture, and livestock farming sectors, improved on-farm prevention strategies, sufficient knowledge about MRSA, VRSA, and VRE transmission routes to the food chain, and determination of high-risk sources of food contamination and the prevalence of mentioned microorganisms in plant- and animal-originated foods are highly needed specially in hot spot regions.
Authors declare that there is no conflict of interest in this work.
The authors confirm contribution to the paper as follows: study conception and design: A.A. and M.H.; data collection: R.R. and B.K.; interpretation of results: S.GH., S.M., and A.A.; writing the manuscripts: A.A., S.M; meta-analysis performance: MA.S.
There was no funding support for this review.
Conflicts of Interest
None.
Received: 2022/05/12 | Accepted: 2022/12/23 | ePublished: 2023/03/30
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |