BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1684-en.html

 , Mohammad Soleimani
, Mohammad Soleimani 

 2, Seyyed Hossein Mousavi3
2, Seyyed Hossein Mousavi3 
 , Mohammad Aminianfar4
, Mohammad Aminianfar4 
 , Rohollah Mirjani5
, Rohollah Mirjani5 
 , Mehran Khoshfetrat3
, Mehran Khoshfetrat3 
 , Monireh Kamali6
, Monireh Kamali6 

2- Department of Microbiology, Faculty of Medicine, AJA University of Medical Sciences, Tehran, Iran , Soleimanidor@yahoo.com
3- Department of Cardiology, School of Medicine, AJA University of Medical Sciences, Tehran, Iran
4- Department of Infectious Disease, School of Aerospace and Subaquatic Medicine, Beasat Hospital, AJA University of Medical Sciences, Tehran, Iran
5- Department of Genetics and Advanced Medical Technology, Faculty of Medicine, AJA University of Medical Sciences, Tehran, Iran
6- Cardiovascular Medical and Research Center, Iran University of Medical Sciences, Tehran, Iran
Forced gram-negative and intracellular bacteria Coxiella burnetii are the most common pathogens of Query Fever disease in humans and Coxiellosis disease in animals (1). Q fever is a zoonotic and endemic disease with a global spread (1). Coxiella burnetii is an immobile bacterium that completes its life cycle in the phagosome of infected cells. It has a cell membrane similar to other gram-negative bacteria but does not take on warm staining. Q fever has been associated with animals (2), but it has been suggested that clinical disease causes little or no disease in domestic animals. In animals, the main clinical manifestations of this disease are abortions in cattle, sheep, goats, cats, and other animals (3, 4).
Coxiella burnetii has been identified as a bacterial pathogen as one of the 13 world-priority zoonoses with an infectious dose of less than one bacterium (5). This pathogen has several notable properties, including proliferating in phagolysosomes such as single-nucleus phagocytic vacuoles, biphasic life cycle, and lipopolysaccharide phase change (6). During the two-phase growth cycle, C. burnetii develops highly resistant spore-like structures known as small cell types (SCVs) that provide long-term environmental stability. However, other fluids and body secretions are also infectious and may facilitate vertical and sexual transmission (4, 7). Spore-like structures are resistant to environmental stressors such as drying or sunlight. Bacteria survive long in soil or other dry matter under adverse conditions (8).
In humans, Q fever may present with a wide range of symptoms, acute, chronic, asymptomatic, or mild (5). More serious manifestations include meningoencephalitis, myocarditis, endocarditis, and chronic fatigue syndrome in patients with persistent infection (6). In humans, the disease has a variety of clinical signs and dangerous complications. Human infection with the disease occurs mainly through inhalation of contaminated animal products or direct contact with infected animals, drinking unpasteurized milk, and consuming other dairy products contaminated with C. burnetii. Thus, Q fever is a common human-animal disease that affects a wide range of hosts.
Since C. burnetii is highly contagious, its resistance to heat and drying in the outdoor environment is high (7). It can also be sprayed and transmitted by inhalation and can cause disease in susceptible individuals (8, 9). Therefore, from the point of view of biological warfare experts, it is a suitable weapon. In the classification of biological weapons, it is classified as a second group of organisms, all of which are usually easily propagated (10, 11). Mobile resistant bacteria are deadly, and many researchers have tried to fight these microorganisms worldwide (12). One way to deal with these bacteria and their infections is to diagnose them quickly and quickly.
Various ways of disease transmission and due to the lack of accurate, comprehensive, and reliable study and the lack of statistics on the percentage of C. burnetii infection in Iranian hospitals on the one hand and the very high resistance of this bacterium in the environment, on the other hand, the study of contamination A hospital is important for this bacterium. Preliminary studies on C. burnetii are mostly based on serological tests to determine the presence of antibodies against C. burnetii in the patient's blood. Rarely and in a limited number of studies polymerase chain reaction (PCR) has been used to detect this pathogenic microorganism (13). Meanwhile, serological methods are used as the first line of diagnosis (14, 15).
Other methods have been used in various studies to diagnose this bacterium. For example, indirect immunofluorescence (IFA) is the standard gold method of bacterial diagnosis. In this method, a fourfold increase in antibody titer in two sera taken from the patient in two different periods must be confirmed for the patient to be positive (16). Due to the complexity and time-consuming method, molecular methods such as PCR can be mentioned, which have the advantages of early detection and high speed of presentation of results (17, 18). PCR is a sensitive and rapid molecular method to detect various pathogens (19, 20).
With a molecular diagnosis, cases of C. burnetii infection can be determined, and the extent of infection can be determined, and as a result, sufficient and documented information based on scientific research and molecular testing can be provided to be effective in the decisions of health managers. This study aimed to molecularly detect C. burnetii bacteria causing infective endocarditis in military hospitals.
DNA Extraction
At this stage, 100 samples of negatively cultured endocarditis were collected from selected military hospitals. DNA extraction from C. burnetii bacteria was performed with the DNP extraction kit of Sinaclon Company (Iran, DNP; lot No. EX6071) and according to the kit instructions. The concentration and quality of the obtained DNA were measured using a nanodrop device.
Primer Design
For PCR, the primers were designed using Gene runner software (version 2) and CLC sequence viewer (version 6) and made by Sinaclone. In order to finalize the designed primers and ensure their specificity, the primers were blasted using the information registered on the NCBI website, and their accuracy was confirmed.
Amplification and Isolation of IS1111 C. burnetii Gene
For each PCR, 25 μL of reaction mixture consisting of 12.5 μL of Master mixture, 1 μL of each primer (cox-F: 5'- GATCATTTGGGCGCTTTTAAC - 3 'and cox-R: 5'- CATCAGCCCTCATTGTTTCG concentration -3') 10 picomoles, 5 μL of template DNA and 5.5 μL of water were prepared by PCR. Two samples were prepared for the positive control (C. burneii; 126 bp) and negative (instead of DNA equivalent water was added) (21).
PCR according to the method (21) under initial denaturation at 94°C for 3 minutes, followed by 35 denaturation cycles at 94°C for 30 seconds, annealing at 52°C for 35 seconds, and expansion at 72°C for 35 seconds, the final extension was performed at 72°C for 5 minutes. Then electrophoresis was performed on 2% agarose gel, and the gel was stained with Red Safe (22).
Positive Control Synthesis
Due to the fact that a positive sample was not available for bacteria, positive control for these bacteria was synthesized based on the sequence of primers and prepared in PUC 18 vector.
GATCATTTGGGCGCTTTTAACACGCCAAGAAACGTATCGCTGTGGCGCCTAAACACCGCCGTGGGTATAAAAAAAGAATTAACAAAAGGAGACACACCAACCGAGTTCGAAACAATGAGGGCTGATG
Feature Determination
In order to evaluate the specificity of primers and determine the specificity of the method, PCR reaction with the conditions mentioned above was performed on the genome of negative control samples. Also, to ensure the ability of the extracted DNA to be used in the PCR reaction and the absence of inhibitors, the PCR reaction on the DNA of bacteria (Staphylococcus aureus (ATCC 25923), Bacillus subtilis (ATCC 6051), Shigella sonnei (ATCC 9290), Escherichia coli (ATCC 25922), Enterobacter faecalis ATCC 29212) and Pseudomonas aeruginosa (ATCC 27853) were performed.
IS1111 Gene Cloning C. burnetiii
To clone a portion of the IS1111 C. burnetiii amplified gene in plasmid PUC 18, the PCR product was first purified using the Bioneer gel extraction kit protocol (South Korea, Cat No. K3037 AccuPrep® PCR / Gel Purification Kit). Cloning was performed using SinaClon TA Cloning Kit (Iran, SinaClon TA Cloning Kit; Cat No. CL5841) according to the kit instructions and with PUC 18 vector. After ligation, JM107 E. coli susceptible to calcium chloride was used to accept the plasmid containing the gene (23).
After susceptibility of the bacteria to plasmid acceptance, 10 μL of the plasmids containing the gene was added to the microtube containing the susceptible bacterium, and finally, the recombinant plasmid was transferred into the bacterium using heat shock. The bacteria were then placed in a sugar incubator at (150 rpm, 3h, 37° C) for 60 minutes. Transformed bacteria were cultured on LB Broth medium containing Ampicillin antibiotic. The culture medium was then centrifuged at 9000 rpm for 2 minutes. From residual precipitate on plate surface (50 mL) LB agar containing (50 mg/mL) Ampicillin 100L, 100L X-gal (20 mg/mL), 80L IPTG (24 mg/mL) and Tet (10 mg/mL) 100 L, Added and broadcast. The plate was placed in a sugar incubator at 37°C for 16 hours.
The following methods were used to confirm the cloning and transfer of recombinant plasmids into bacteria:
Formation of White Colonies Containing Recombinant Vectors
After culture, the transformed bacteria are seen in the culture medium of blue colonies (colony without recombinant plasmid) and white (colonies containing recombinant plasmid).
For PCR colony, 2 to 3 white colonies from the transformation stage were selected, and PCR reaction was performed using PCR Master mix (Ampliqon; Cat. No.: A190303). The product was then electrophoresed in 1% agarose gel to confirm the presence of the cloned fragment.
Extraction of Recombinant Plasmid
Plasmid extraction was performed after confirmation of the presence of recombinant and inserted plasmids using ViVantis Plasmid Extraction Kit (Malaysia, Plasmid DNA Extraction Kit; Cat No. GF-PL-050 GF-1). In the quantitative evaluation of plasmid extraction, the first 1:10 and 1:100 dilutions were obtained from the plasmid extraction sample. After reading the concentrations of the dilutions prepared by the nanodrop device, the final concentration of the extracted plasmid was obtained in a volume of 100 microliters.
Sensitivity and Detection Limit
To determine the sensitivity and limit of detection (LOD), the minimum number of copies of the desired fragment that can create a clear band in the PCR reaction was calculated. For this purpose, successive double dilutions (1-10 to 10-10) of PUC18 plasmid with specific concentrations were prepared, and PCR was performed using them. Its products were electrophoresed on 1% agarose gel. The last dilution in which the PCR product bonds will be the LOD that can be obtained with CN = MN / LD, the least number of traceable copies.
DNA Extraction
Pure DNA was obtained using a nanodrop device for samples from patients with endocarditis (Figure 1). By examining the result of quantitative DNA analysis, its amount was calculated between 1.69 and 1.8.
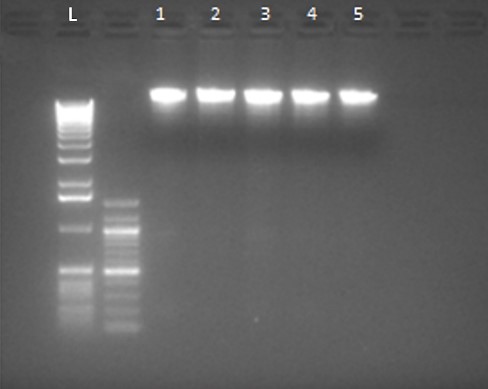
Figure 1. Extracted DNA sample (L: DNA Ladder and 1-5: Purified DNA)
Based on the collected samples, endocarditis was examined for the presence of C. burnetii in these patients. Still, out of 60 samples, no positive samples were reported for any of these factors.
Specification
In order to investigate the specificity for each pair of primers, PCR reaction with the conditions related to each gene was performed on the genome of the negative control bacteria extracted. Still, no band was observed after electrophoresis of PCR products related to each gene on the agarose gel (Figure 2). This indicates that the designed primers attach only to the target bacterium.
Figure 2. Specification for C. burnetiid. Negative control bacteria from left to right L size marker, C. burnetii, Staphylococcus aureus, Bacillus subtilis, Shigella sonnei, Escherichia coli, Enterobacter faecalis, Pseudomonas aeruginosa, Klebsiella pneumoniae, respectively
Determine LOD
The last dilution of PUC18 plasmid for C. burnetii with an initial concentration of 700 ng/µL, which formed a detectable band on the gel, was calculated to be 7-10, and the minimum number of detectable copies in a 25 μL PCR reaction equal to 22 copies.
Fever is an emerging and emerging zoonosis in many countries, including Iran. Determining the prevalence of infection and risk factors makes the importance of infection for health officials, and the necessary facilities and equipment for control and prevention as well as research priorities are identified (24, 25). The results of studies show that the one-step PCR method for the diagnosis of C. burnetii is not sensitive enough, and it is recommended to use the nested PCR method. This method has high speed, accuracy, specificity, and sensitivity compared to classical methods (26, 27).
In recent years, many studies have been performed to identify C. burnetii, which has been identified in various infected samples. Still, few cases of this bacterium have been studied in hospital samples (28, 29). In the study conducted by Kirkan et al., in 2011. Of 296 samples, 12 (4%) were positive (30). Astobiza et al., in a study of 178 samples in Spain, showed that 119 of the 178 suspected cases were positive for the presence of anti-C. burnetii antibodies (26, 31). Dosti et al. conducted a study on 130 camel blood samples. In this study, 14 cases (10.76%) of camel blood samples by PCR were positive for the presence of C. burnetii (32). In 2012, Giuraniks et al. Conducted a study in Hungary on 215 samples, 7 of which 67% were positive for C. burnetii (33).
Literak had not seen any human infection with Coxsackie in the Czech Republic and Slovakia in the five years leading up to 1996 (39). However, in his report, Hirai reports a positive increase in cases of human infection with C. burnetii in Japan in recent years (39). In a 1999 scientific study in Japan, his colleagues noted that 33% of those tested were infected with the microorganism (40). Guo et al. also detected coxsackie infection in 1998 in 3.4% of sheep farmers in North Dakota (USA) (41).
The bacterium has QPRH1 plasmids in acute infection, and the disease develops as atypical pneumonia. QPRH1 is related to the acute form of the disease; this group is sensitive to chloramphenicol and tetracycline. The bacterium contains QPRS plasmids in chronic infection, and the disease develops as chronic endocarditis years later. This group of plasmids is resistant to chloramphenicol and tetracycline. The third group without plasmids is involved in causing chronic disease. Endocarditis is the main clinical manifestation of chronic Q fever and can lead to death in more than 65% of patients if left untreated (42-44). Therefore, the diagnosis of genes in acute or chronic forms of the disease is very important in developing drug resistance, which is one of the major medical problems today (45). The absence of Query Fever at the time of the study and in the past, or the absence of specific clinical symptoms related to Query Fever in the past, are consistent with the results of this study (39).
In this study, C. burnetii was not detected in any blood samples. However, C. burnetii has a wide and varied host spectrum in the Epizootic and Enzootic foci. Comparing the results of our study with the previous research works of researchers in Iran and other countries also reveals obvious differences or relative similarities. Each pathogen's enzootic (native animal) cycles, including C. burnetii, are different in reservoir hosts in different regions. Accordingly, statistics show significant differences in the prevalence of coxsackie in different geographical areas.
Based on the initial serological evidence obtained, it seems that Q fever is currently present in Iran, and due to the few studies available, the disease is not considered or is confused with other febrile diseases such as malaria or influenza. Due to the zoonotic nature of the disease and the potential contamination of host animals, it is necessary for the veterinary agencies and organizations, and subdivisions of the Ministry of Health to interact extensively and closely to identify reservoirs and prevent and control the disease. Given the health and economic importance of the disease in humans and livestock, it is recommended that ruminants in the disease reservoir be identified after slaughter and that at-risk individuals such as veterinarians, ranchers, and slaughterhouse workers be vaccinated.
None.
None.
Conflicts of Interest
The authors declared no conflict of interest.
Received: 2022/02/27 | Accepted: 2022/05/15 | ePublished: 2022/09/9
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |






