BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1521-en.html
2- Department of Microbiology, Razi Vaccine and Serum Research Institute, Agricultural Research, Education and Extension Organization (AREEO), Karaj, Iran ,
3- Department of Research and Development, Razi Vaccine and Serum Research Institute, Agricultural Research, Education and Extension Organization (AREEO), Karaj, Iran
Leptospirosis is one of the most important diseases transmitted from livestock to humans caused by the bacterium Leptospira (1). The disease has a wide geographical spread worldwide, not only in third world countries but also in developed countries. According to the World Health Organization (WHO) report, this is the second disease transmitted from livestock to humans (2, 3). It is most prevalent in temperate and tropical regions, especially in areas with high rainfall (4, 5). Leptospirosis has also been observed in areas with neutral or slightly alkaline pH, such as northern Iran (6). Unfortunately, for various reasons, this disease is not diagnosed correctly. In addition, pathogenic bacteria in soil and water with the right pH and temperature can survive for a long time alongside non-pathogenic leptospira. For this reason, a rapid and accurate diagnosis of the disease and the separation of pathogenic species from non-pathogenic is one of the most important measures that must be taken to prevent, control and properly treat the disease (7, 8).
Different PCR and hybridization methods are among the molecular detection methods that each is used according to the characteristics in different fields of classification and identification of various Leptospira strains (9, 10). Molecular methods such as PCR are of great significance for accurately detecting infectious serovars. Due to the zoonotic nature of this bacterium and because the treatment will be effective only in the first days of the disease, it is, therefore, crucial to correctly and quickly diagnose Leptospirosis (11, 12). Bacterial membrane proteins are the basis of the link between the bacterium and the host and are highly stable among pathogenic species (13, 14). Accordingly, many attempts have been made to identify the properties of leptospiral membrane protein components. Identification of outer membrane proteins (OMPs) of pathogenic Leptospira has recently emerged as a central research topic for Leptospira. The leptospiral membrane proteins such as LipL21, LipL41, LipL32, OmpL1, LigB and LigA can be appropriate targets for identifying these bacteria (15-17). Among these membrane proteins, OmpL1 has a high antigenic heterogeneity among pathogenic Leptospira species, but LipL32 and LipL41 membrane proteins are structurally more stable and have very few alterations (18). Among these proteins, Loa22 has been identified in pathogenic Leptospira. Various studies have suggested that Loa22 is expressed during infection, can be detected in the patients' serum, and elicit an immune response in patients. Loa22 is also a surface-exposed protein, which provides partial protection in hamsters (19, 20). These observations indicate a possible association of this protein with bacterial virulence (21). Accordingly, the given work aimed to investigate the presence of gene encoding Loa22 in pathogenic and non-pathogenic Leptospira serovars as a molecular marker to identify pathogenic Leptospira.
Leptospira Serovars
The present study was done on 23 pathogenic leptospira serovars and two non-pathogenic leptospira serovars. These serovars were prepared from the Reference Laboratory for Leptospira, Department of Microbiology, Razi Vaccine, and Serum Research Institute, Karaj, Iran (Table 1).
Table 1. Serovars tested in research
| RTCC | Serovar | Serogroup | Number | RTCC | Serovar | Serogroup | Number |
| 2828 | Patoc | Semanerga | 14 | 2802 | Autumnalis | Autumnalis | 1 |
| 2829 | Pomona | Pomona | 15 | 2805 | Canicola | Canicola | 2 |
| 2830 | Autumnalis | Autumnalis | 16 | 2808 | Grippotyphosa | Grippotyphosa | 3 |
| 2831 | Malaysia | Malaysia | 17 | 2810 | Hardjo | Serjoe | 4 |
| 2835 | Pyrogenes | Pyrogenes | 18 | 2812 | Icterohaemorrhagiae | Icterohaemorrhagiae | 5 |
| 2836 | Canicola | Canicola | 19 | 2815 | Pomona | Pomona | 6 |
| 2837 | Icterohaemorrhagiae | Icterohaemorrhagiae | 20 | 2817 | Serjoe | Serjoe | 7 |
| 2838 | Ballum | Ballum | 21 | 2819 | Patoc | Semanerga | 8 |
| 2839 | Javanica | Javanica | 22 | 2821 | Hardjo | Serjoe | 9 |
| 2840 | Australis | Australis | 23 | 2822 | Pomona | Pomona | 10 |
| 2841 | Laitype lanylokowii | Laitype lanylokowii | 24 | 2823 | Icterohaemorrhagiae | Icterohaemorrhagiae | 11 |
| 2843 | Serjo hardjobovis | Serjo hardjobovis | 25 | 2824 | Canicola | Canicola | 12 |
| 2825 | Grippotyphosa | Grippotyphosa | 13 |
The Culture of Leptospira Serovars
Bacteria were cultured in Leptospira-specific EMJH (Difco) culture medium with a pH value of 7.5±0.5. Incubation was performed at 28°C for 5-7 days. To study the growth and density of bacteria, their proper motility, and the absence of secondary contamination, slides were prepared from the cultures and explored under dark field microscopy.
DNA extraction
First, cell deposition was prepared to extract DNA. To this end, 70-80 ml of fresh bacterial culture was poured into a sterile Falcon tube and centrifuged at 4°C under 17000 ×g for 20 minutes. Then, 1 ml was harvested from the remaining cell deposition and centrifuged at 17000 ×g for 20 minutes. The remaining cell deposition was applied to extract DNA. The standard extraction method of phenol: chloroform: isoamyl alcohol was employed for DNA extraction. After extraction, 30-40 μL of 1x TE buffer was added to each microtube based on the amount of DNA deposition, and the microtubes were placed in a thermoblock at 60°C for 60 minutes to dissolve the DNA completely. The extracted DNA was stored at -20°C for subsequent testing. The quantity and quality of the extracted DNA were evaluated by spectrophotometry and 1% agarose gel electrophoresis, respectively.
Polymerase Chain Reaction (PCR)
Specific primers designed in this study for loa22 gene were used to identify pathogenic Leptospira serovars and differentiate them from non-pathogenic serovars. The primers could amplify a 671-bp fragment of the loa22 gene (Table 2). Thus, 6 μL of 2X Master Mix with 1 μL of each primer at a concentration of 10 pmol and 1 μL of DNA sample at a concentration of 100 ng reached a volume of 12 μL with distilled water and placed in a thermocycler. The temperature cycle consisted of an initial denaturation cycle at 94°C for 5 min with 35 cycles, including denaturation at 94°C for 1 min, annealing at 62°C for 1 min, and extension at 72°C for 1 min. A final extension step was performed at 72°C for 10 min. To observe the bands obtained by PCR, after the end of thermocycling, 1% agarose gel electrophoresis was carried out and the resulting bands were observed by the gel doc system.
Table 2. Sequence and specifications of loa22 gene-specific primers
| GC% | Tm (˚C) | Fragment Length (bp) | Sequence (5' → 3') | Primers |
| 43.48 | 58.87 | 671 | CGGCCTTTTGAAAGATCGAATTG | Forward primer |
| 47.62 | 57.87 | 671 | ACACTCTGATACCAAACCCCT | Reverse primer |
Determining the Sensitivity of Primers
The extracted and quantified DNA of L. Grippotyphosa (RTCC 2808) was used to reach the minimum limit of detection of DNA by PCR using specific primers. Thus, the DNA sample was diluted from 100 ng/µL to 0.0001 pg/µL. Finally, 1 μL of each dilution was poured into each microtube and PCR was performed with specific primers.
The Characterization of Primers
It is essential to determine the specificity of the loa22 gene primer for pathogenic samples of this bacterium. In this section, PCR was performed with specific primers on DNA extracted from two pathogenic species of Leptospira, including L. interrogans Pomona (RTCC 2815) and L. interrogans Australis (RTCC 2840), a saprophytic sample including L. biflexa (RTCC 2819), and some bacteria including Shigella sonnei (RTCC 1870), E. coli (RTCC 2325), Salmonella enteritidis (RTCC 1621) and Citrobacter freundii (RTCC 1096).
Determining the Presence of loa22 Gene in the Studied Serovars
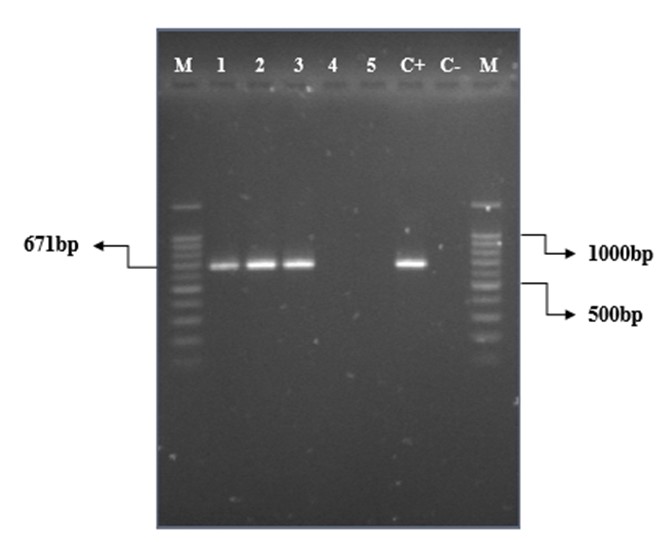
PCR with specific primers of loa22 gene was performed on purified DNA of 23 pathogenic Leptospira serovars and two saprophytic Leptospira serovars. As shown in Figure 1, a 671-bp fragment was found only in pathogenic Leptospira serovars. In contrast, this gene was not observed in non-pathogenic serovars, indicating the presence of the loa22 gene for pathogenic Leptospira.
Figure 1. PCR amplification results of loa22 gene in Leptospira serovars (M: 100-bp molecular marker (Bio-Rad), 1: L. autumnalis (RTCC 280 2), 2: L. grippotyphosa (RTCC 2808), 3: L. pomona (RTCC 2815), 4: L. biflexa (RTCC 2819), 5: L. biflexa (RTCC 2828), C+: L. canicola (RTCC 2805) as the positive control, C-: negative control)
Determining the Sensitivity
As can be seen in Figure 2, the PCR sensitivity with selective primers was high and positive results were observed up to a dilution of 0.01 pg. These results indicate that if DNA is diluted up to 0.01 pg, it can be identified and amplified by this test along with specific primers.

Figure 2. Results of electrophoresis to determine sensitivity to different dilutions of purified DNA of pathogenic Leptospira serovars: L. serovar Grippotyphosa (RTCC 2808) (1 and 13: 100-bp molecular marker (Bio-Rad), 2: 100 ng, 3: 10 ng, 4: 1 ng, 5: 100 pg, 6: 10 pg, 7: 1 pg, 8: 0.1 pg, 9: 0.01 pg, 10: 0.001 pg, 11: 0.0001 pg, 12: Negative control)
The Characterization of Primers
As shown in Figure 3, the gene encoding the Loa22 protein was present in pathogenic leptospira serovars. Still, it was not found in the non-pathogenic L. biflexa serovar (RTCC 2819) as well as in four other bacteria, indicating the specificity of this primer for pathogenic serovars so that it is not observed in non-pathogenic serovars and other bacteria.

Figure 3. Results of electrophoresis for PCR products on 1% agarose gel to characterize the primers (1 and 11: 100-bp molecular marker (Bio-Rad), 2: L. interrogans serovar Pomona (RTCC 2815), 3: L. interrogans serovar Australis (RTCC 2840), 4: L. biflexa (RTCC 2819), 5: Shigella sonnei (RTCC 1870), 6: E. coli (RTCC 2325), 7: Salmonella enteritidis (RTCC 1621), 8: Citrobacter freundii (RTCC 1096), 9: negative control, 10: positive control L. interrogans serovar Canicola (RTCC 2805))
In recent years, various serological and molecular research on Leptospira have been conducted in Iran, such as determining the genetic pattern of Leptospira serovars used in Leptospirosis vaccine in Iran using VNTR (MLVA), molecular detection of Leptospira using the 16S rRNA genom fragment, cloning of genes expressing LipL32, OmpL1, LigB proteins of Leptospira interrogans in Razi Vaccine and Serum Research Institute of Karaj (22-24). However, there is still no standard method for rapid and accurate disease diagnosis in medical diagnostic laboratories and health centers (25). The Loa22 protein could also be a new candidate for molecular identification of Leptospira serovars to protect against leptospiral infection. However, further research is needed to confirm the importance of Loa22 in pathogenicity as well as protective activity (26).
The findings of this study showed that the PCR method could be used to distinguish the genus Leptospira from other bacteria as well as pathogenic species from non-pathogenic ones. Since no studies have been performed on the loa22 gene of pathogenic and native Leptospira serovars in Iran so far, it is necessary to study and identify this gene. PCR is a rapid, sensitive, and specific method for diagnosing leptospiral infections, especially in the early stages of the disease (27). Because Loa22 protein has been identified in pathogenic Leptospira species but is not present in non-pathogenic ones, the PCR test on the loa22 gene is very useful for detecting pathogenic Leptospira species in clinical practices.
The present study recruited specific and designed loa22 primers to identify pathogenic serovars from non-pathogenic ones. The primer used for the gene encoding Loa22 protein was capable of amplifying the 671-bp fragment. Based on the results of PCR in this study, the 671-bp fragment was observed in 23 pathogenic Leptospira serovars but not in two saprophytic serovars. Hence, the gene encoding the Loa22 protein was present in 23 pathogenic Leptospira serovars but not in the two non-pathogenic Leptospira serovars. This indicates that this gene is expressed only in the pathogenic serovars studied. In the present study, the specificity of primers designed for loa22 gene was also investigated, the results of which demonstrated that these primers were specific for pathogenic Leptospira serovars and other bacteria and non-pathogenic Leptospira serovars showed no amplification with these primers. The sensitivity of the primers was about 0.01 pg of DNA, indicating their high sensitivity.
Varadarajan et al. (2015) in India reported an upregulation of Loa22 during clinical Leptospirosis in dogs compared to other OMPs. This study examined 70 blood samples from dogs referred for clinical Leptospirosis. In addition, 12 pathogenic Leptospira reference serogroups were used. By PCR, the presence of the loa22 gene in all 12 pathogenic serogroups and lipL32 and ligB genes were reported in 11 and 7 positive serogroups, respectively. According to PCR findings on dog blood samples, the loa22 gene was detected in 15 samples, but the other two genes were not detected in any of the samples. Finally, the results of this study, in line with the present study's findings, showed that the loa22 virulence gene could act as a diagnostic marker for Leptospirosis (28).
Meenambigai et al. (2016) in India evaluated the sensitivity of the Loa22 virulence gene to multiplex PCR to diagnose Leptospirosis in dogs. They employed multiplex PCR to target the three genes of loa22, lipL32, and lipL21 to overcome the limitations of MAT and isolation methods. This team applied 12 pathogenic reference serogroups of Leptospira as well as 40 dog blood samples. The results revealed that out of 12 references pathogenic Leptospira serogroups, the Loa22, and LipL32 genes were detected in all 12 serovars with 100% sensitivity and the LipL21 gene in 9 serovars with 75% sensitivity. Among 40 dog samples, 12 out of 40 dog samples were positive for the loa22 gene, 5 samples were positive for the lipL32 gene, and only 3 samples were positive for the lipL21 gene. The results of this study demonstrate the sensitivity of the diagnosis of the loa22 virulence gene in canine Leptospirosis (29).
Koizumi and Watanabe (2003) in Japan investigated the presence of Loa22 protein among 17 pathogenic and non-pathogenic Leptospira serovars using immunoblotting with anti-Loa22 serum. They found that a positive signal reacting with anti-Loa22 serum could be detected in the same region as Loa22 in all pathogenic serovars. At the same time, no detectable levels of protein were observed in non-pathogenic serovars such as L. biflexa and L. meeri. Therefore, a strong association was observed between virulence and the presence of Loa22, indicating the involvement of this protein in the pathogenesis of Leptospira. Finally, this study reported that Loa22 was detected in pathogenic Leptospira but not in non-pathogenic Leptospira (26). A single article published by Haake et al. in 2015 states that a homolog of the loa22 gene with 56% sequence homology is present in L. biflexa (30). In the future, this gene can be used in cloning and expression of a recombinant antigen, which can be used in the preparation of an effective and efficient recombinant vaccine, as well as in serological diagnostic kits such as ELISA. All of these require further testing in this area.
Our results were consistent with the findings of other researchers regarding the presence of the loa22 gene in pathogenic Leptospira and its absence in non-pathogenic serovars, indicating the possible role of this gene in the pathogenesis of Leptospira. According to the results obtained in this study, it can be concluded that the loa22 gene can be recruited in molecular diagnosis to distinguish between two pathogenic and non-pathogenic Leptospira serovars using PCR technique.
The authors are very grateful to the microbiology department of the Razi Vaccine and Serum Research Institute.
The financial resources of this research have been provided from Grant with the financial number 12-18-18-106-96045-961023 of Razi Vaccine and Serum Research Institute.
Conflicts of Interest
There is no conflict of interest between the authors.
Received: 2021/10/18 | Accepted: 2022/05/28 | ePublished: 2022/08/8
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |