BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1302-en.html
2- Professor of Department of Medical Microbiology, Faculty of Medicine, and Applied Microbiology Research Center, System Biology and Poisonings Institute, Baqiyatallah University of Medical Sciences, Tehran, Iran ,
3- Professor of Medical Microbiology, Faculty of Medicine and Health Management Research Center, Baqiyatallah University of Medical Sciences, Tehran, Iran
4- Professor of Medical Microbiology, Applied Microbiology Research Center, System Biology and Poisonings Institute, Baqiyatallah University of Medical Sciences, Tehran, Iran
5- Assistant professor of Medical Microbiology, Faculty of Medicine, and Applied Microbiology Research Center, System Biology and Poisonings Institute, Baqiyatallah University of Medical Sciences, Tehran, Iran
Streptococcus pneumoniae (S. pneumoniae) is an important human pathogen and a leading cause of morbidity and mortality worldwide (1). Invasive and noninvasive infectious diseases caused by S. pneum-oniae strains such as meningitis, sepsis, bacteremia, pneumonia, sinusitis, acute otitis media in young children, elderly, and patients with immunodeficiency are increased (2,3). In previous studies, the frequency of pneumococcal meningitis in elderly patients was reported more than 35% (4-6).
The main virulence factor of the S. pneumoniae is capsular polysaccharide which protects the bacteria against phagocytosis and plays an important role in colonization the bacterium on the upper airway (7). Thus, expression of polysaccharide capsule genes is essential for survival in against of the superficial imm-une, and enhances the virulence and pathogenicity.
Currently, the strategy for the prevention of life-threatening pneumococcal disease, is based on the immunogenicity of polysaccharide capsule and the production of capsule base vaccines and existing vac-cines (PCV7, PCV10, PCV13 and PPSV23) which made from capsule antigens (8). However, the causes such as the spread of serogroups, and emergence of new serotypes and serotype diversity (9) have reduced the efficacy of vaccines. Therefore, the use of vaccine in all cases could not be a good solution for preventing pneumococcal disease and its morbidity and morta-lity. In addition, the development of antibiotic resist-ance has increased the therapeutic problems (10). This bacterium has been resistant to many common anti-biotics, therefore, since the production of polysac-charide capsules are introduced as the most important pathogenicity and virulence factor (11, 12). Several study on capsular synthesis mechanisms and also inhi-bit expression the involve genes were more consid-ered (11).
Based on available data, in most serotypes, the capsule locus is located in the same region between the genes dexB and aliA locuses. The first four genes of this locus are conserved and named as: cpsA, cpsB, cpsC and cpsD, and thus, to determine the level of expression of the capsular genes locus are very important, (13, 14). It is well known that the cpsB and cpsD genes, plays major role in regulating the capsular synthesis and production (15)
It is reported that; Xylitol is a five-carbon sugar alcohol (16, 17) and have affected the bacterial growth and blocked the adhesion property of S. pneumoniae (18,19). Research results have shown that Xylitol can prevent acute otitis media infections in children (20). Some research results revealed that psaA gene produces a 37-kDa protein called Pneumococcal sur-face adhesin A (21), which acts as a superficial protein for binding of S. pneumoniae to mucosal cells (22, 23). As a result, it is reported that the psaA gene is also involved in the disease. Since, all pathogenic S. pneu-monia have been shown to have this gene (24). Thus, the purpose of this study was assayed the effect of different concentration of Xylitol on the expression of the cpsD and psaA genes. If the genes were inhibited, probably Xylitol can be used as complementary treat-ment with no any side effects against the S. pneum-oniae superficial infection hopefully.
Bacterial strain
This investigation was carried out during the year 2019 in Baqiyatallah university complex laboratories. A reference strain of S. pneumoniae (ATCC 6305, pur-chase from Baharafshan Laboratory, Tehran, Iran), and twenty S. pneumoniae clinical isolates which pre-viously were identified and their serotypes were reported (25, 26) was used. In separately manner, the lyophilized bacteria vials in aseptic conditions were dissolved in 1 ml BHI- broth media (Merck Co, Cat No.110493). Then 50µL of the suspension was trans-ferred into Blood agar (Merck Co, Cat No.110886) which supplemented by 5% sheep blood and incubated at 37°C in a 5% CO2 atmosphere for 24 hours. After overnight incubation, bacterial colonies were harvested and transferred into 3 ml of BHI broth media containing 0.2% glucose and supplemented with 10%(vol/vol) FBS (Fetal bovine serum) and incubated at 37°C in a 5%CO2 atmosphere until the OD650 had been reached equal 0.5 McFarland appro-ximately for 5 hours. For each strain the bacterial suspension as the same of standard 0.5 McFarland (107 CFU/mL) were considered. In order to confirma-mtic assay for all strain of the S. pneumoniae, a stand-ard diagnostic procedure was carried out based on previously report (27, 28).
However, for each reaction the 300 µL of bacterial suspension was added to 3 ml of test media that were prepared by adding various sugar alcohol or sugar concentrations (Table-1). The test tubes were incub-ated at 37°C in 5% CO2 atmosphere for about 3 hours. In order to obtain proper concentration cells and the desired RNA level, the contents of each test medium was centrifuged in 4°C for 3 min at 12000g that super-natant containing the BHI broth was discarded and the resulting sediment was used to RNA extraction.
Sugars stock solution preparation and sterilization
In this research, the 40% stock solution of each sugar was prepared separately. Tested sugars were sterilized by filtration (0.22 µm pore-sized filter; Milli-pore.). Afterward certain amount was added to relat-ed test tubes. The Xylitol concentration used in this st-udy were chosen on the basis of a previous study (19).
RNA Extraction
The kit of the TRIzol® Reagent solution (Cat No.-15596-026; Ambion; USA) was used in order to extract RNA. It should be noted that in the whole process of extraction of RNA, it is necessary to note that due to the abundance of the RNase enzyme in the envir-onment and the sensitivity of RNA, moreover to use filter micropipette tips and RNase free micro tubes, and the whole process is done alongside the ice.
Table 1. Media and ingredients used for bacterial growth and RNA extraction. The volume and different concentrations of Xylitol, Fructose and Glucose were prepared from the stoke solution.
| Total volume | Sugar | FBS | BHI broth | Media |
| 3000 µL | 375 µL | 200 µL | 2425 µL | BHI broth + 5% Xylitol |
| 3000 µL | 375 µL Xylitol and 375µL fructose |
200 µL | 2050 µL | BHI broth + 5% fructose + 5% Xylitol |
| 3000 µL | - | 200 µL | 2800 µL | BHI broth Glucose -free (Control) |
Based on the kit procedure, the RNA was extracted and stored at minus 70 ° degree centigrade. The pro-bable DNA contamination, the extracted total RNA was subjected to treat by Dnase1 enzyme (Cat No. PR891627; GENEALL; South Korea). As follows: 1 µL of DNase I, 10x enzyme was added to 1 μg extracted RNA in a RNase free micro tubes. 1 µL of buffer 10 x was added and the final volume with DEPC reached 10 µL and incubated for 30 minutes at 37°C. The reaction was completed by adding 1 µL of 50 mM EDTA solution and incubation at 65°C. in order to analysis of the quality and quantity of the extracted RNA, the absor-bance of 1.9 to 2 at 280/260 ratio revealed the purity of extracted total RNA by using Nano drop device (ND-1000; Thermo). In order to determine the quanti-fication extracted specimens, the concentration was calculated according the absorbance at 260 nm.
To evaluate the quality of extracted RNAs, samples were run in agarose gel 1% (100 volts). The extracted RNA has shown a good quality, it will have two bands in gel, including 16srRNA and 23srRNA band. After th-at, all RNAs samples were rapidly converted to cDNA.
cDNA Synthesis
For cDNA synthesize, the PrimeScript™ RT reagent Kit (Cat No. RR037A; TAKARA; Japan) was used. The reaction mixture was prepared according to the instruction of the kit (Table-2). Briefly, this mixture was incubated 15 min at 37℃ for rverse transcription and then for inactivation of reverse transcriptase was incubated 5 secoun at 95℃. To prevent contamination and experimental error until before the Real-Time PCR, wasn’t use from specific primers for each of the genes and there were three total cDNA types.
Table 2. The Reagents and gradients utilized to synthesis cDNA.
| concentration | Amount | Reagent | |
| 1X | 2 µL | 5X Prime Script Buffer | |
| - | 0.5 µL | Prime Script RT Enzyme Mix I | |
| 50 μM | 0.5 µL | Oligo dT Primer | |
| 100 μM | 0.5 µL | Random 6 mers | |
| 454.4 µL/ng | 3.5 µL | Extracted from control media | Total RNA |
| 395.8 µL/ng | Extracted from media containing 5% Xylitol |
||
| Extracted from media containing 5% fructose + 5% Xylitol |
|||
| - | 3 µL | RNase Free DH2O | |
| - | 10 µL | Total volume | |
RT Real Time PCR Performance
For this purpose, specific primers were designed with GenScript software for amplify target genes (cpsB, cpsD and psaA), and to be synthesized were ordered (Macrogen; South Korea) (Table-3).
Briefly, 5 µL of synthesized cDNA was used in Real-Time PCR by applying the Real Q Pluse 2X Master Mix Green low ROX™ with SYBR® Green (Cat No. A323499; Ampliqon; Denmark) and the Exicycler TM 96 Real-Time Quantitative Thermal Block (Cat No. A-2060; BIONEER; Republic of Korea). The therm cycler programs and protocols and gradients reagent were used according the kit instruction.
Table 3. Characteristics of Designed primers for set up the RT Real Time PCR
| Concentration | Amplicon size | Tm(°C) | GC% | Sequence 5’ – 3’ |
Primer | |
| 5 Pmol/µL | 202bp | 58.4°C | 50% | TAGATGACGGTCCCAAGTCA | Forward | cpsB (wzh) |
| 5 Pmol/µL | 56.4°C | 45% | GCGCCATAAGCAATGACTAA | Reverse | ||
| 5 Pmol/µL | 159bp | 58.4°C | 50% | CCAAACCCTACAGCCTTGTT | Forward | cpsD (wze) |
| 5 Pmol/µL | 56.4°C | 45% | TGTTACCAAGATGGACGCAT | Reverse | ||
| 5 Pmol/µL | 254bp | 58.4°C | 50% | GTAGCATGTGCTAGCGGAAA | Forward | psaA |
| 5 Pmol/µL | 54.3°C | 40% | TTTGTAAACCAAGCATTGCC | Reverse | ||
| 5 Pmol/µL | 286bp | 54.3°C | 40% | TTTGGCATCAATCTGTCCAT | Forward | aroE |
| 5 Pmol/µL | 56.4°C | 45% | AAACGGAACGAACAAAGACC | Reverse | ||
However, The Real-Time PCR was carried out in accordance with the instructions of the utilized kit. Real Time PCR was performed two times and every sample was run as triplicates in each time. Finally, the average CT was totally obtained from the 6 collected CTs for each sample.
As it was shown in figure 1, Real-time PCR assay was performed to evaluate the expression of the cpsB, cpsD and psaA genes on total cDNAs related to media containing 5% Xylitol and glucose -free (Control) media. According to obtained results of and by using REST 2009 software, the automatic statistical analysis by REST 2009 software revealed that, the expression of cpsB, cpsD and psaA genes was affected (see part A and B of Figure 1), but only the expression of cpsB gene in expose with 5% Xylitol was reduced signifi-cantly in compared to the glucose free media as Con-trol (P=0.005). However, there was no significant red-uction in the expression of the cpsD and psaA genes.
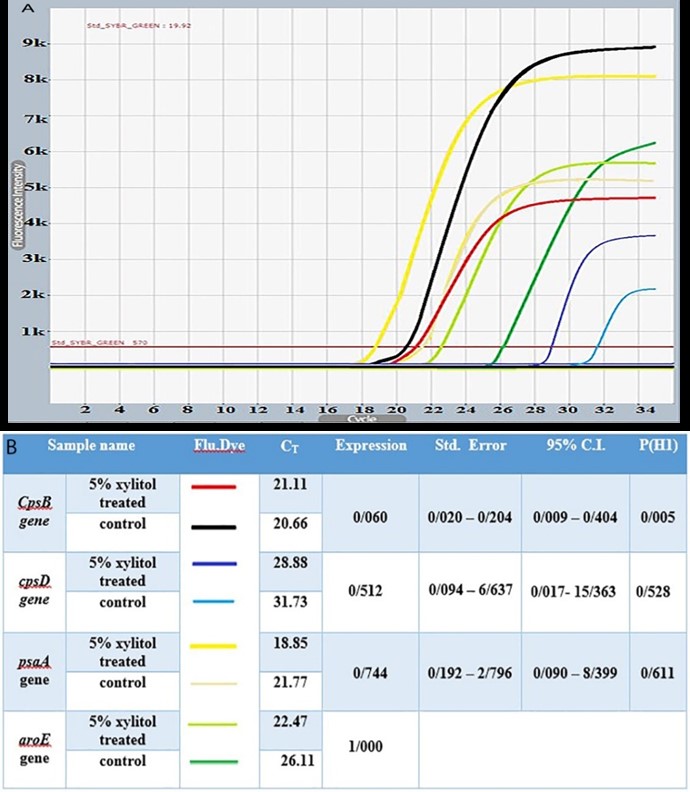
Figure 1. The Cyber green RT Real Time PCR amplification has showed. Part A), the pattern of implications cycle of cpsB, cpsD, and psaA genes in different media in this study. Part B), the results of REST 2009 software statistical analysis has shown.
In the second set up to simultaneously evaluate the effects of Xylitol and fructose; Real-time PCR assay was carried out on total cDNAs related to medias containing 5% Xylitol and 5% fructose + 5% Xylitol. It's just here, Only the expression of cpsB gene was reduced and the expression of the cpsD and psaA genes did not change (Figure-2). This finding also confirmed the results of previous studies that the presence of fructose in the media prevents the inhibition of Xylitol.

Figure 2. Part A) The resulting pattern of replication of cpsB, cpsD, and psaA genes in media containing 5% Xylitol compared to media containing 5% fructose + 5% Xylitol. Part B) The REST 2009 software results statistical analysis has shown.
The analysis of melting curve also proved during the Real-Time PCR, only one specific product was ampli-fied and there is no non-specific product and primer-dimer. More than one peaks show non-specific ampli-fication. Melting curve results of cpsB gene showed that the Tm of the all product of this gene was equal to 82°C and also in the case of cpsD gene, in the analysis of melting curve only one peaks at 82°C was observed.
Emerging a wide range of deadly infections by S. pneumonia (29) as well as the development of antibiotic resistance by this bacterium (30). Diagnosis, treatment and prevention of relative infections are global problem (31). In addition, a vaccine that can cover all pathogen strains is not yet available. There-fore, other ways to control pneumococcal infections have been considered in recent years.
One of the current hypothesis that could possibly prevent the infection from pathogenic strains is focused on the inhibition of some virulence genes.
Since the S. pneumonia several virulence genes that one of the most important of these genes is the polysaccharide capsule regulator genes (as the most important virulence factor) and the psaA gene, which plays a major role in bacterial binding, can be ment-ioned.
Therefore, cpsB, cpsD and psaA genes were invest-igated in this study and as control, the aroE gene, also known as the House Keeping gene, was examined.
A published study results have shown that the fructose could prevent the inhibitory antibacterial action of Xylitol; this may be due to Xylitol entering bacteria through fructose receptors while in the abse-nt of fructose, its inhibitory effect does not occur (19). As the same, our previous study results revealed that the presence of 5% fructose inhibit the anti- pneumo-coccal activity even after 24 hours (32).
Furthermore, our previous study had shown the growth pattern of bacteria based on the recorded ODs by MTT method, and concluded that the best time and concentration of the user sugars. Present study was conducted out to assay the effects of the expression of selected genes, and 3 hours after exposed of bacterial suspension to 5% Xylitol, 5% fructose + 5% Xylitol as a point out were considered. Thus, pertain-ing to the present study, it should be noted that in less than 3 hours, there is not enough time to evaluate the inhibition of the expression of genes and in more than 3 hours, the growth rate of bacterial cells will vary in different media, Therefore, the time point of 3 hours, which was equivalent to MIC concentration of the 5% Xylitol, was chosen to assessment the inhibition of the expression of the genes based on the previous study finding (32).
However, in this study based on our previous expe-rience, we used 3 hours as a time point out for invest-igating the assay of inhibitory effects of utilized sugars against the selected gens expression. While, the Kuor-ola's research were implemented on standard strain ATCC 49619, the expression of the only cpsB gene was monitored 2 hours after exposure to 5% Xylitol and The 16SrRNA gene was also selected as the reference gene (20). In addition, the Kuorola's resea-rch were focused on one target gene expression, but in this study, we assayed four target gene. However, pertain-ing to the one target gene the results were the same.
According to the results of the present study, the relative expression of cpsB gene in exposure to 5% Xylitol was significantly reduced ccompared with 5% glucose and control media. The outstanding of the current research was that, the results of the real-time PCR were analyzed by using REST 2009 software in order to determine the expression of cpsB, cpsD and psaA genes of bacteria strains and also reference ATCC 6305 strain in 5% Xylitol, 5% fructose + 5% Xylitol and control medias. Also the aroE gene was used as the reference gene. According to the REST 2009 software analysis (see attached Tables in Figures 1 and 2), although the expression of target genes was associa-ted with a decrease, but the decrease in cpsB gene expression was significant renege in compared to control (P=0.005).
By comparing these results with the results of Kuorola studies, it was observed that in the same experimental conditions, including the same volume of BHI- Broth, FBS and similar concentration of Xylitol, the expression level of cpsB gene in this study was reduced significantly; the differences may be due to differences in standard bacterial strains, reference gene type, exposure duration, different quality of RNA extraction kits, cDNA synthesis, and Real-Time PCR protocols.
In recent years, the health benefits of using Xylitol have received much attention; not only act as an oral hygiene effects but also it stimulates the immune system. In addition, a research results have shown that the short-term consumption of Xylitol may decreasing the release of pro-inflammatory cytokines and the counts of Streptococcus mutans. Although, a plenty of research has been reported on the benefits of Xylitol, its antibacterial mechanism of action is unknown. Thus, in the present study, three genes encoding the capsular- making proteins were invest-igated. The main finding was that, after 3 hours’ expo-sure to 5% Xylitol, the relative expression of cpsB gene was reduced about 97.8 % compared with control media (P=0.005). This is an outstanding result, beca-use of it may be very important and suggest the possi-bility of clinical use of this sugar.
The authors would like to thank the Deputy of Clinical Development Medical Center of Baqiyatallah Hospital for their support.
Conflicts of Interest
The authors declared no conflict of interest.
Received: 2021/03/14 | Accepted: 2021/07/15 | ePublished: 2021/08/16
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |








