BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1267-en.html
2- Department of Microbiology, School of Medicine, Iran University of Medical Sciences, Tehran, Iran ,
Acinetobacter baumannii is an aerobic non-fermenting gram-negative coccobacillus belonging to the family Moraxellaceae. This opportunistic bact-erium is usually isolated from the hospital envir-onment and hospitalized patients. Acinetobacters, especially A. baumannii, are a part of the natural skin flora, which can localize the mouth cavity, pharynx, and tonsil. This point is important in terms of epidemiology and nosocomial infections [1].
Some of the most important infections caused by this bacterium include endocarditis, meningitis, pneumonia, bacteremia, and urinary infection [2]. Over 43% of healthy people have this organism on the surface of their body [3]. A. baumannii is considered one of the remarkable problems in nosocomial infections in the world due to antibiotic resistance. The clinical significance of this bacterium is seriously threatening, especially during the last 11 years because of acquiring antibiotic resistance genes [4].
This pathogen produces biofilm-associated (Bap) protein by the bap gene that starts biofilm formation following A. baumannii attachment to cellular sur-faces. Outer membrane protein A (OmpA), expressed by the ompA gene, is an adhesion protein, which plays an important role in biofilm formation on the surface of epithelial cells. Exopolysaccharide (Esp) is coded by the espA gene in high levels on cell surfaces and protects the cell against different types of envir-onmental damage. Esp protein is involved in the aggregation and biofilm formation by this bacterium. Rapee Thummeepak et al. in 2016 reported 84.4%, 48%, and 30.2% of ompA, bap, and espA in 255 clinical samples, respectively [5].
The Csu protein generates a secretory system, inclu-deing chaperone and usher, which are needed for the production of pilli and formation of biofilm on surfa-ces. The expression of this protein is controlled by a regulatory system of two components named Bfms-/BfmR that entails a kinase-sensor coded by bfmS and a response regulator coded by bfmR. This system is applied for bacterial biofilm formation on polysty-rene surfaces. It has been demonstrated in previous studies that inactivating the bfmR gene leads to the cessation of pilli production, adhesion, and biofilm formation on plastic surfaces. The frequency of this gene has been reported to be high in clinical isolates. Thummeepak et al. found a frequency of 84% for this gene [6].
Other virulence factors encompass iron acquisition system (basD), phospholipase D (slpD), surface antigen protein A (surA), and penicillin-binding protein G (bpbG) [7, 8]. Therefore, the present study aimed to identify nine diverse virulence genes in A. baumannii isolates isolated from patients with pneumonia hospitalized in the Intensive Care Unit (ICU) of Imam Khomeini Hospital, Tehran, Iran.
Isolation and Identification of Isolates
A total of 50 isolates of A. baumannii were randomly selected from the microorganisms collection of the Iran University of Medical Sciences. These bacteria were selected from the clinical samples of the respiratory system of patients who referred to Imam Khomeini Hospital, Tehran, Iran during 2018-2019 [9]. Biochemical tests, including oxidase, catalase, anaero-bic oxidation-fermentation (OF), aerobic OF, triple sugar iron (TSI), indole, motility, culture on Simmons Citrate agar, Voges-Proskauer, methyl red, culture on MacConkey agar, and lactose test (Merck, Germany) were performed to ensure correct colony selection. Finally, 50 A. baumannii isolates were identified and confirmed [10].
Primers Designing
The target sites for A. baumannii primers were chosen as nine specific genes, namely bap, ompA, basD, espA, bfmR, lpD, csuA, pbpG, and surA. Following the determination of target sites for primer designing, databases of each bacterium were searched on NCBI. The Oligo 2, Oligoanalyzer, and Gene Runner softwares were applied to assess the annealing and Tm temperatures of primers (Table 1). Next, blasting with human, fungal, viral, and microorganism samples was performed with NCBI databases for the designed primers.
Table 1. Sequences of primers used for identifying specific genes in A. baumannii isolates
| Gene | Primer Sequence | Product Size | Reference |
| OmpA | F: GCTGGTGTTGGTGCTTTCTG R:TCGGTTGATCCCAAGCGAAA |
490 | This study |
| Bap | F:TGAAAGTGGCTGCCAGTGAT R:TCTGCGTCAGCGTCACTATC |
223 | This study |
| EspA | F:CAGCTTTAGGTCTTGCGGGA R:TGCTAAACCGAAGTCGCCAA |
392 | This study |
| PbpG | F:TGGATGCGCAAACAGGTGAA R:GGTCGGTGTGGTTGAGAACT |
467 | This study |
| BasD | F: TGCTGTTCGTTCTTTTGGCG R:GTTGAGTTTGAGCGCCGATG |
517 | This study |
| PlD | F: GCTGTGGCTTTGACAGGTTG R TAGCGCAAACGGTGTTGTTG |
695 | This study |
| BfmR | F:ACCGATGGTAACCGTGCAAT R:TCTGCACCCATTTCCAGACC |
194 | This study |
| SurA | F:GATGCGATTGCACCTGGAAC R: TTGACGTGCCATACGCTCTT |
822 | This study |
| CsuA | F: TGGTGAAGCTACCACAGGTT R:ACGACTACCATCATGGGCTG |
322 | This study |
DNA Extraction from A. baumannii Isolates
First, colonies of all A. baumannii isolates were separately cultured in microtubes containing 2 mL of Tryptic Soy Broth (TSB) medium and were incubated at 37°C for 24 h, which followed by the extraction of genomic DNA from colonies by the boiling method.
First, bacterial suspension was centrifuged at 10000 rpm for 10 min after 24 h incubation. Afterwards, 1 mL of the supernatant was discarded and 1 mL distilled water was added to the bacterial precipitate. This procedure was performed twice. Next, 200 µL of TE sterile buffer (pH: 8) was added to the bacterial precipitate. Microtubes containing precipitate and TE buffer were boiled at 100°C for 10 min and were immediately transferred to the temperature of -20°C for 5 min. The process of boiling and cooling was repeated three times. Afterwards, the microtubes that contained bacterial precipitation were centrifuged at 14000 rpm. Finally, the supernatant containing genomic DNA was collected in a sterile microtube for polymerase chain reaction (PCR). In order to determine the purity of extracted DNA, electroph-oresis and biophotometer were carried out.
Multiplex PCR
PCR was performed using Taq DNA Polymerase Master Mix (Amplicon, Denmark) which consists of MgCl2, dNTPs, Taq polymerase, and buffer. The final volume of each reaction for completing multiplex PCR was 25 µL containing 12 µL Master Mix for all reactions. Moreover, 0.3 µL at the concentration of 10 pM was poured in a microtube from each forward and reverse primer of ompA gene with 1 µL of both primers for bap gene. In addition, 0.5 µL of both reverse and forward primers of the three genes csuA, plD, and basD were poured into another tube.
The amounts of 1, 0.5, and 0.5 µL of the reverse and forward primers of pbpG, bfmR, surA genes were added to the third microtube, respectively. A volume of 0.5 µL of each primer of the espA gene was poured into another tube. The espA gene was evaluated separately and not as multiplex. The final concentration for each primer was 100 µmol. After that, 1 µL DNA at the concentration of 200 ng/L and 1 µL double-distilled water free of any nuclease was added to each reaction mixture to reach the intended final volume.
Multiplex PCR was carried out as follow: 1) primary denaturation for 5 min at 95°C, 2) 34 denaturation cycles of 40 sec at 95°C, 3) annealing for 45 sec (annealing temperature was determined as 59°C for all nine genes, 4) polymerization for 45 sec at 72°C, and 5) final polymerization for 5 min at 72°C. Afterwards, PCR products were electrophoresed on 1% agarose gel containing safe stains and were visualized by gel documentation. The PCR products were finally sent to Pishgam Co. (Iran) for sequencing.
Statistical Analysis
Statistical analysis was performed using the SPSS software version 22 (IBM, Chicago, Ill., USA). The relationship between antibiotic resistance and the frequency of ompA, bap, bfmR, espA, basD, surA, pbpG, csuA, and plD was assessed by the t-test.
Identification of A. baumannii
Microbiologic and biochemical tests were perfor-med for the definite determination of 50 A. baumannii isolates collected from the respiratory system of patients. In the laboratory, all 50 isolates were cultured on MacConkey agar and blood agar and were incubated at 37°C for 24 h. After that, the presence of Gram-negative coccobacilli A. baumannii was confirm-ed by a microscope. Next, biochemical tests, such as IMViC, urease, TSI, OF, MR, VP, SIM, catalase, oxidase, and growth at 37°C and 42°C were conducted to diagnose diverse species of Acinetobacter.
Isolates that were lactose negative, non-motile, oxidase negative, catalase positive, indole negative, pigmentation negative, urease positive, citrate pos-itive, H2S negative, and MR negative were confirmed. The results of microbiologic and biochemical tests demonstrated that all 50 samples were A. baumanni and stored at -70°C in Peptone Water medium containing 30% glycerol.
Results of PCR for Specific Genes
All samples identified as A. baumanni by bioch-emical tests were tested by multiplex PCR for ompA, bap, bfmR, plD, basD, espA, csuA, pbpG, and surA genes. The results of multiplex PCR revealed that the genes bfmR, bap, ompA, and csuA involved in biofilm formation had the frequencies 100%, 98%, 96%, and 98%, respectively. Gene espA was found to have the lowest frequency of 10% among the isolates. The lengths of products are demonstrated in Table 1.
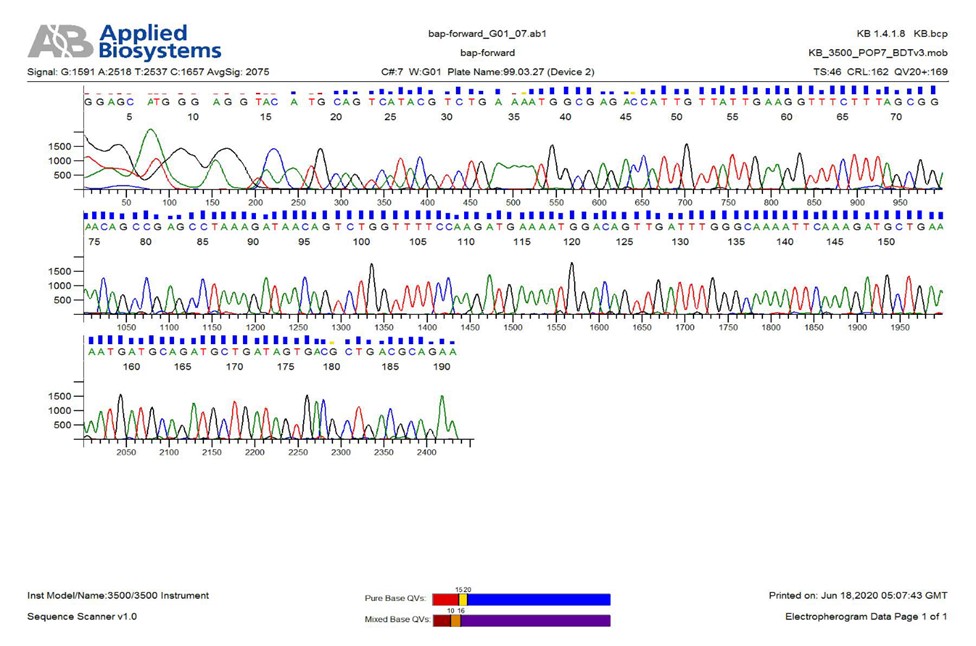
According to the findings of previous studies, the bap gene is the most important gene for biofilm formation [5]. The frequency of this gene was very high in the current study (98%) and only one isolate lacked this gene. The frequency of other investigated genes in the present study was 98%, 96%, 98%, and 100% for plD, basD, surA, and pbpG, respectively (P<0.05) (Figures 1-5). For all negative specimens, PCR was repeated twice as a single gene. The frequency of genes basD, bfmR, bap, ompA, plD, csuA, pbpG, and surA was 82% (Table 2). The PCR products were finally sent to Pishgam Co. for sequencing. The findings of sequencing showed a similarity of over 80% in isolates (P<0.05) (Figure 6).

Figure 1. Frequency of studied genes in A. baumannii isolated from patients
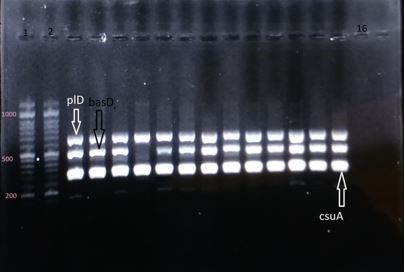
Figure 2. Electrophoresis gel of the PCR products of genes plD (the segment of 695 base pair), basD (the segment of 517 base pair), csuA (the segment of 322 base pair); columns 1 and 2: DNA marker of 1000 base pair; column 16: negative control

Figure 3. Electrophoresis gel of the PCR products of genes surA (the segment of 822 base pair), pbpG (the segment of 467 base pair), bfmR (the segment of 194 base pair); column 1: DNA marker of 1000 base pair; column 15: negative control
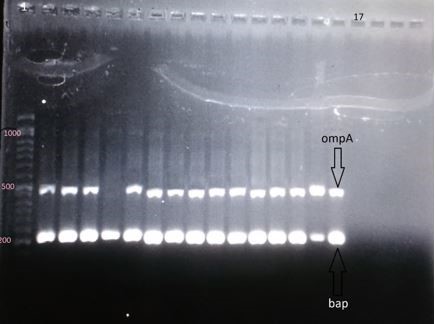
Figure 4. Electrophoresis gel of the PCR products of genes ompA (the segment of 490 base pair), bap (the segment of 223 base pair); column 1: DNA marker of 1000 base pair; column 17: negative control
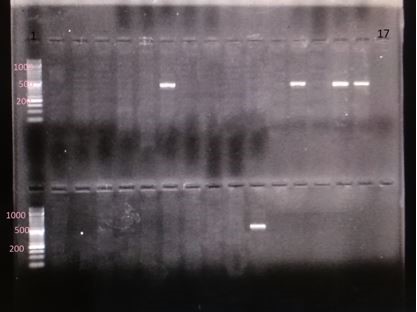
Figure 5. Electrophoresis gel of the PCR products of genes espA (the segment of 392 base pair); column 1: DNA marker of 1000 base pair; column 17: negative control
Table 2. Pattern of pathogen factors in A. baumannii isolates isolated from the respiratory system
| ompA | bap | espA | basD | plD | bfmR | surA | CsuA | pbpG | Pattern | Percentage | Number of isolates |
| + | + | - | + | + | + | + | + | + | P1 | 82% | 41 |
| - | - | - | + | + | + | + | + | + | P2 | 2% | 1 |
| + | + | - | - | - | + | - | + | + | P3 | 2% | 1 |
| + | + | + | + | + | + | + | + | + | P4 | 8% | 4 |
| + | + | + | + | + | + | + | - | + | P5 | 2% | 1 |
| + | + | - | - | + | + | + | + | + | P6 | 2% | 1 |
| - | + | - | + | + | + | + | + | + | P7 | 2% | 1 |
Figure 6. A sample of bap gene sequencing
During the last decade, A. baumannii has been identified as the most important nosocomial pathogen throughout the world. It is partly due to the living ability of this microorganism in the hospital environment and obtaining resistance mechanism leading to acute infections, especially in severely ill patients. Although extensive information exists concerning the mechanisms of antibiotic resistance in this microorganism, it cannot be stopped yet [11].
Although the severity of A. baumannii infection is directly related to the pathogen factors, the frequency of these factors in clinical samples of A. baumannii is less reported [12]. Therefore, the infection caused by this agent and the immune response of the host to these infections is still ambiguous. In the present study, the frequency of nine genes of virulence factors were evaluated in the clinical samples of A. baumannii isolated from the respiratory system of patients. Biofilm is considered as an important factor in clinical specimens of A. baumannii. The extension and thickness of biofilm structures and intracellular attachments are tightly related to bap family proteins, which are coded by the bap gene [13, 14].
Our results concerning the frequency of pathogen genes were highly similar to the findings of Chaoliu et al. in China. In the mentioned investigation, the frequency of the bap gene in the clinical samples of A. baumannii was reported as 87.5%. Furthermore, Fallah et al. in 2019 found the frequency of 92% in 100 clinical isolates in Iran [15, 16].
The ompA is a protein of the outer membrane that induces biofilm formation on the epithelial cells of humans. This protein is coded by ompA [5]. In Iran, Bardbari et al. (2017) and Zeighami et al. (2019) reported the frequency percentage of this gene as 100% in clinical isolates of A. baumannii isolated from respiratory samples and 81% in the isolates related to other clinical specimens, respectively [17, 18]. Their results were highly similar to the findings of the current investigation (96%). Although the ompA gene has a high frequency, previous studies have shown that biofilm formation is more associated with the bap gene, compared to the ompA gene [19].
The csuA gene belongs to the csu gene operon, which consists of six genes (csuA/BABCDS) and codes the chaperone-usher secretory system as a very important part of pilli generation and biofilm structure formation on non-living surfaces [20]. The bfmR gene as the regulatory part of the two-component regulating system had a frequency of 100%. The pres-ence of this gene in A. baumannii isolates is essential for pilli generation and biofilm formation on plastic surfaces [6]. Thummeepak et al. in a study in Thailand (2016) on A. baumanni clinical samples reported a frequency of 84% for the latter gene [5]. These findings revealed that most of the clinical isolates of A. baumannii had the potential for biofilm forming. The frequency of the espA gene in the present study (10%) was lower than the other genes that contribute to biofilm formation, including csu, bfmR, espA, ompA, and bap.
Thummeepak et al. and Kim et al. (2019) observed the frequencies of 30% and 50% for the mentioned gene in 255 clinical samples and 181 specimens isolated from blood culture in South Korea, respectively [5, 21]. Two other genes investigated in the current study were plD and surA, both of which are serum resistance factors [7, 22]. Chaoliu et al. reported the frequency of 95% for surA and 92% for plD, which is consistent with our results [15].
The basD was another gene with a frequency of 96% in the current study. Chaloliu et al. and Kim et al. revealed it to be 92% and 98%, respectively [15, 21]. Finally, the pbpG gene that codes PBPs had a frequency of 100% in A. baumannii isolates. Although PBPs are the receptors of beta-lactam antibiotics, are known as one of the pathogen factors in A. baumannii isolates and account for bacterial survival in human serum and the soft tissues of animal models [20].
The findings of some investigations had a very low similarity with our results. The latter discrepancy could be attributed to the type of studied samples, study time, and the type of used antibiotic disk. On the other hand, these bacteria benefit from diverse mechanisms for pathogenicity, scape host immune system, and antibiotic resistance. Numerous studies have been performed in Iran on the epidemiologic characteristics and drug resistance pattern of A. baumannii isolates. However, it is necessary to consider the role of this bacterium as a potentially dangerous factor in nosocomial infections.
The genes involved in the pathogenicity of A. baumannii have a high frequency and the nosocomial prevalence of this bacterium has elevated. Therefore, large-scale studies and protection programs, such as infection control in the ICU seem to be necessary. Limiting the consumption of antibiotics, development and changing of hygiene plans for contaminated facilities, and classifying patients in whom this bacterium has colonized are among the beneficial approaches for controlling the spread of A. baumannii.
The present paper is related to the Master thesis of the first author.
The current study has financially been supported by the Research Deputy of Iran University of Medical Sciences with the grant code of 98-4-4-16126. The authors would like to thank the latter deputy.
Conflicts of Interest
The authors declared no conflict of interest.
Received: 2021/01/4 | Accepted: 2021/02/13 | ePublished: 2021/06/28
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |