BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1262-en.html
2- Department of Microbiology, School of Biological Sciences, Islamic Azad University, North Tehran Branch, Tehran, Iran ,
3- Department of Medical Mycology and Parasitology, Division of Molecular Biology, School of Public Health, Tehran University of Medical Sciences, Tehran, Iran
Introduction
The World Food Organization (FAO) and its 191 member states have introduced June 1st as the World Milk Day since 2001, in line with its commitment to protect the public access to adequate and high-quality food for healthy living. The organization has urged all governmental and non-governmental organizations to inform people of the milk importance as the most comprehensive and indispensable food for all groups on this day (1).
Milk as a complete food source contains nutrients, minerals and vitamins needed by the body and as one of the most important products of the agricultural sector, it is of great importance. About 85% of human milk is consumed from cow's milk (1).
Milk is a complete meal for human consumption and its health condition is very effective in human health. Since milk is a rich and complex environment of various nutritional compounds; it can also be useful for other microorganisms, including fungi and bacteria. This nutrient can be contain various compounds including proteins, carbohydrates, fats, vitamins and minerals (2).
Since raw milk is used in the various dairy industries, including yogurt, cheese, dough, etc., the composition of the microbial flora is remarkable. Although most attention has been paid to bacterial flora, but also fungal flora, since some yeast can be used as a starter in the dairy industry, or as a probiotic, it is important to examine them, and it can be helpful to identify and raise the level of health of the milk to raise the level of public health (3).
Determining the primary quality of the fungal flora of the milk can be identified by identifying secondary contamination that occurs during transportation and in environmental conditions. In order to investigate these factors, we made the necessary precision at all stages, including livestock hygiene, tank health, and hygiene devices. Yeast and molds can also be an important microbial population in raw milk that is affected by the conditions of animal, physiology, the weather, season and nutrition (3).
Milk can be a source of different non-starter fungi, although microflora of different dairy industries such as dough, yogurt, and cheese has been investigated, but identification of the primary source can be better for us (4). In a 2010 study in China, 11 yeast species were identified in milk (5). Few studies have been done on the cow's milk variety. Identification of the yeast used in the cheese industry as a starter is helpful as it is also effective in the flavor and taste of the cheese (4).
But until this study, the presence of fungal flora in Iranian milk has not been reported. Most research is on milk samples contaminated with mycotoxins.
The results of this study determined the fungal flora of milk in the samples and can be used in somatic studies of tuberculosis to identify related animal diseases such as mastitis in herds. Also, the reported type of yeast can affect the price of milk, which is directly related to its quality in factories, and can be classified based on the amount of fungal flora reported.
The purpose of this study was identification and molecular characterization of the fungal-microflora in raw cow's milk. This study focused on illustrating the diversity of yeast species in cow's raw milk from industrial farms of Tehran province and Alborz, Iran.
Materials and Methods
Selection of Farms
In this Cross-sectional study, random farms were selected from Tehran and Alborz provinces, Iran. The samples were collected from 14 farms and examined during March 2016 to March 2017. Milk samples were collected by random sampling every month from the milk collection tanks and transferred by sterile Falcon tubes (0-4°C) to the laboratory. Tanks containing milk for more than 1 h were excluded from the study. Samples were collected immediately after sterilization of the last livestock in the farm in a sterile Falcon tube. It did not reach the time of one hour and so on. This study was sampled in four seasons as shown below.
Isolation of Yeast Flora
First, the samples were cultured for preparation. Milk samples were cultured on Yeast Extract Agar (YEA) with antibiotics (penicillin & streptomycin 1%) and incubated at 35°C for 48-48 h. Then grown-up colonies were stained by the lactophenol cotton blue (LCB) for yeast shape. Evolution and were passaged on Sabourate Dexterose Agar (SDA) with antibiotics and placed at 35°C for 48-48 hours.
Diagnosis of Yeasts using CHROM Agar Medium
Some types of yeasts are specifically identified by creating colored colonies on the chromogenic agar medium. Accordingly, we incubated yeasts on this medium at 30°C for 48 hours and then examined them.
Identification of Yeast Species using RFLP-PCR:
Yeast DNA extraction was performed according to a procedure previously described by using phenol-chloroform technique (6). DNA concentration and purity were determined by Nanodrop (Therm-oscientifics, USA).
PCR analysis
To amplify the internal spacer region ITS1-5.8S-ITS-2 of the yeast rDNA genes, two universal primers, ITS-1: 5"TCCGTAGGTGAACCTGCGG3" and ITS-4: 5"TCCTC-CGCTTATGATATGC3" were used (7). PCR analysis of genomic DNA was performed according to the standard protocol using synthetic oligonucleotide primers. Briefly, 2.5 μL of each primer (OD 20, 10 pmol) was added to a volume of 45 μL containing: 10X PCR buffer + MgCl2, 10 mM dNTP mix, 1 unit/μL of Taq DNA polymerase and template (genomic DNA) (Amplicon, Denmark). Thermal cycle reaction was carried out by a thermal cycler (Peqlab. Germany). It was composed of a pre-denaturation at 94°C for 5 minutes followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at 58°C for 45 seconds, and a final extension step at 72°C for 75 seconds. PCR products were analyzed by electrophoresis through a 1% agarose under ultraviolet Trans illumination.
RFLP
The MSPI (HpaII) enzyme was used to digest PCR products (7). Briefly, 10 µL of PCR product, 1.5 µL of enzyme, 3µL Tongue buffers, and 18 µL nuclease –free water were mixed and the mixture was incubated for 1.5 hours at 37ºC. Restricted fragments were then revealed by electrophoresis through a 2% agarose and the obtained results were analyzed by compare with previously reported data.
PCR Product Sequencing:
The PCR fragments from yeast samples which could not be identified by RFLP method, was forwarded for a sequence analysis. The obtained results were then investigated for identifying in BLAST (NCBI, NIH)
Statistical Analysis
Excel 2016 software (Microsoft, USA) was used for data analysis. The data was entered into Excel and sor-ted by collection time. The average number of total yeasts during different seasons was calculated using Excel software and its standard deviation was deter-mined.
Results
Identification of Yeast
In this study, 262 milk samples were collected from 14 farms. The number of samples collected in Spring, Summer Autumn, and Winter were 55, 79, 74 and 54, respectively. The microbial analysis of milk samples were performed with a focus on normal yeast flora in milk. The total number of isolates for each genus and species from farms was included in Table 1. The yeast samples studied microscopically and then cultured on Chrom-agar medium; the obtained results were analyzed based on the pigments produced and shown in Figure 1.
We did our research in four seasons, and the frequency and type of yeast were shown in Figures 2-5. Among the seasons, the highest levels of yeast were found in the Autumn (Figure 6).Mean and standard deviation number of yeasts by different seasons were shown in Figure7. The most striking feature of this chart is that based on the somatic cell count, which is generally considered to be a measure of microbial load of milk, and is more commonly used for cows' mastitis, as we can see, the number of infected samples were more in Spring.
Table 1. Isolated yeast from milk samples.
| Genus | Total | Species | Total | Number of farms |
|---|---|---|---|---|
| Candida | 72 | glabra | 2 | 1 |
| globosa | 33 | 9 | ||
| inconspicua | 16 | 8 | ||
| tropicalis | 6 | 4 | ||
| Parapsilosis | 10 | 7 | ||
| Utilis | 1 | 1 | ||
| pesudoglobosa. | 3 | 3 | ||
| Galactomyces | 4 | Candidum | 3 | 3 |
| Geotrichum | 2 | 1 | ||
| Geotrichum | 12 | candidum | 12 | 6 |
| Kluyveromyces | 64 | marxianus | 64 | 12 |
| Cryptococcus | 1 | amylolentus | 1 | 1 |
| Cutaneotrichosporon | 11 | curvatus | 10 | 4 |
| Smithiae | 1 | 1 | ||
| Magnusiomyces | 15 | capitatus | 15 | 8 |
| Pichia | 90 | jadinii | 19 | 9 |
| Cactophila | 3 | 1 | ||
| Fermentans | 10 | 5 | ||
| Kudriavzevii | 57 | 12 | ||
| Heedi | 1 | 1 | ||
| saprochaete | 3 | clavata | 3 | 2 |
| sporopachydermia | 1 | lactativora | 1 | 1 |
| Tricosporon | 31 | asahii | 24 | 10 |
| Montovideence | 6 | 5 | ||
| Lactis | 1 | 1 | ||
| Trapelia | 1 | elacista | 1 | 1 |
| Wickeihameilla | 15 | pararugosa | 14 | 9 |
| Sorbophila | 1 | 1 | ||
| Rhodotoroula | 1 | millaginosa | 1 | 1 |
| Meyerozyma | 1 | guilliermondi | 1 | 1 |
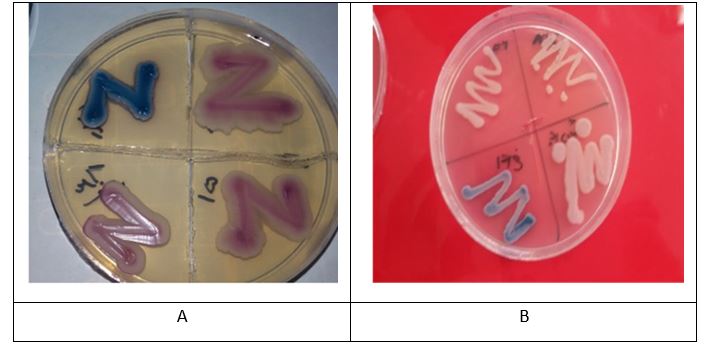
Figure 1. Chrom agar medium
According to the pigments produced, using Chrom agar medium culture, the color of the fungus colony has been determined. Figure A shows the fungus Candida tropicalis and image B shows the fungus Candida Krusei

Figure 2. In this season, the levels of Kluyveromyces marxianus and Pichia kudriavzevii are almost the same.
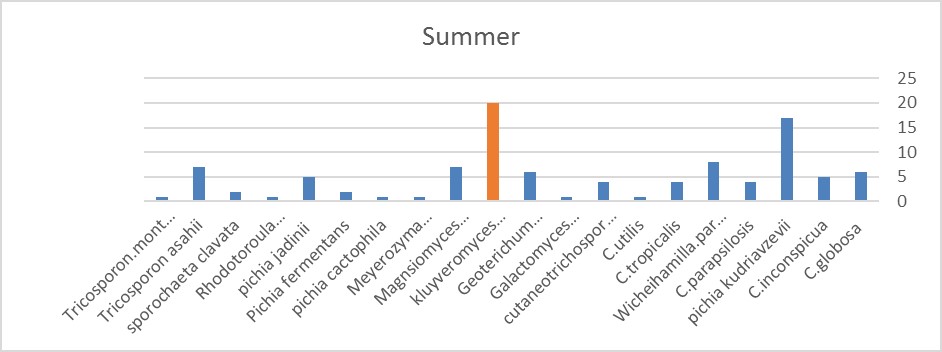
Figure 3. In this season, Kluyveromyces marxianus is more than the others.
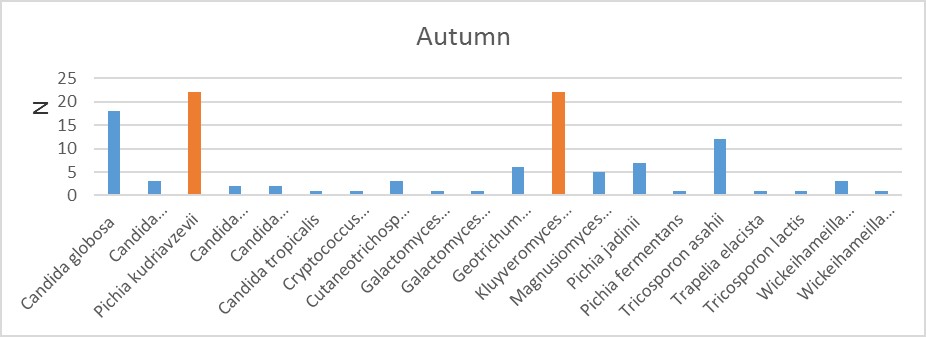
Figure 4. In this season, Kluyveromyces marxianus and Pichia kudriavzevii are the same and more than others.
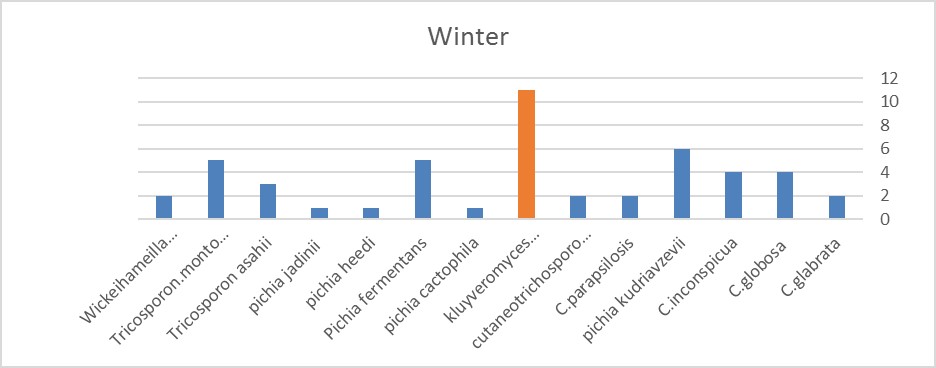
Figure 5. In this season, Kluyveromyces marxianus is more than the others

Figure 6. The yeast in different seasons
.jpg)
Figure 7. Mean (and standard deviation) number of yeasts by different seasons
Molecular Identification of Yeast:
Using the PCR-RFLP method based on the length of the digested parts, the yeasts were identified. The results were indicated in Figures 8 and 9.
Sequencing
Identifying all yeasts method was not possible by PCR-RFLP. Therefore, the nucleotide sequencing method was used to obtain the exact result based on the results obtained and compared with the gene bank.
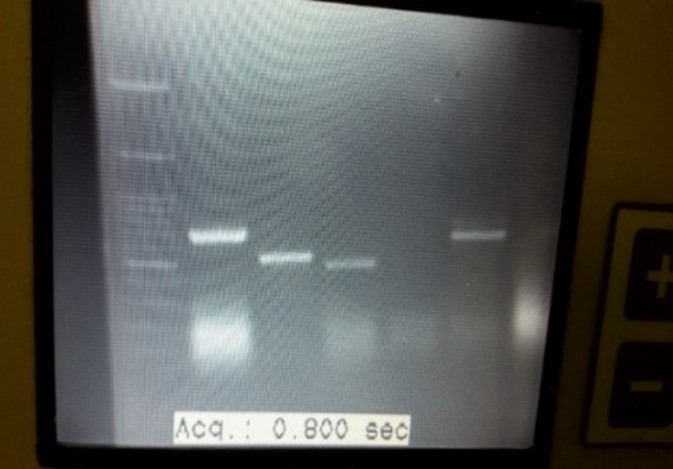
Figure 8. PCR product of Candida Spp.: The length of size marker bands (Lane1) are: 2000-1500-1000-900-800-700-600-500-400-300-200-100 bp (from top of bottom), Lane2-6 are Candida spp.
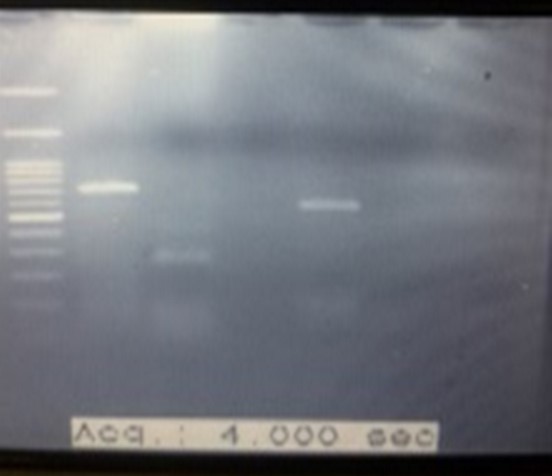
Figure 9. Digestion of PCR Product with MSPI: The length of size marker bands (Lane1) are: 2000-1500-1000-900-800-700-600-500-400-300-200-100bp (from top of bottom), Lane2: kluyveromyces marxianus Lane3: Candida tropicalis Lane5: Candida parapsilisis
Discussion
Milk health is important as a source of food and precursor fermentation product. The interest in drinking raw milk of cattle is growing in some societies, and many people believe that it is important to consume it for their health. Raw milk is a perfect food, however, it can be microbiologically hazardous for consumers. Food is needed for life, but this role can be overshadowed due to the fact that they sometimes cause illness instead of an effect on growth. All foods can be contaminated physically, chemically, and biologically. Milk contributes to both food sources and consumer health (8).
In Europe, raw milk is sold directly in some countries, such as Germany, France, the Netherlands, Belgium, Denmark, Italy, and Ireland (9).
Milk has been found to be a good food source for many types of microorganisms due to its diverse nutrient ingredients that support microorganisms (3). It is also a good source for some probiotics such as Saccharomyces cerevisiae (10).
Yeast play an important role in the fermentation of dairy industries due to lipolytic and proteolytic activity and atopic dermatitis with raw milk in the early years of life (3). Some yeasts produce pathogenic microorga-nisms and inhibit the production of inhibitors (11)
Understanding the microorganisms of Eukaryote in addition to increasing the knowledge of their classi-fication is important because identification of fungi have been considered in terms of the production of aflatoxin and its effects on human health (9, 12, 13). Also, yeasts can be a cause for mastitis in cattle (9). Also, due to the type of microorganisms present in the milk, the taste and flavor of the products derived from milk will be different (14). Therefore, their identifica-tion is very critical and important.
Yeasts such as Candida, Saccharomyces, and Kuyve-romyses have been observed in milk. Yeasts are the main components of the microbiota of fermented mil-ks. In some dairy products, yeasts use lactic acid, whi-ch leads to a decrease in pH, which promotes the seco-ndary growth of pH-sensitive microbiota. The growth of bacteria is also triggered by amino acids and vitam-ins produced from yeasts. Yeasts display a variety of metabolic capabilities, such as proteolytic and lipolytic activity, that can play a significant role in taste and aroma (11).
In 2008, Karmen Godič Torkar in Sloan checked the quality of the milk and reported the presence of yeast and molds with 95% yeast and 51.5% Geoterichum, while our percentage of yeast and Geoterichum contained 81.3% and 3/7%, respectively. Their exam-ination was done only in the winter and summer, but our review was done in four consecutive seasons (15).
Karine Lavoie et al. studied on fungi flora of raw milk and cheese in 2012, which was similar to our research in terms of Candida, Cryptococcus, Galactomyces, Pichia, Rhodotorula, and Tricosporon kluyveromyces detection (4). In a study in China in 2010, conducted by Chen et al., 11 species of yeasts were identified. Information on the fungal diversity of raw milk is low (5). In 2013, in Italy, Simona Panelli et al. studied the variation of fungal microflora in raw milk at Alpine altitudes, based on the height of the grasslands, using srRNA 26, its, and the beta-tubulin gene. Their study showed that increase in height of the grasslands is related with increase of fungal diversity. Yeast dominant microorganism and Kluyveromyces marxi-anus was the most abundant (12) which was similar to results of the present study. In the same year, in other areas of Italy, cheese products were also studied, and yeasts were identified (16). Mostly, research has been done on milk products such as cheese and raw milk. In 2013, Nataša Golić, discovered yeasts in the study of soft fresh cheese in Serbia, which included Kuyverom-yces, Debaromyces, Saccharomyces, Candida, and To-rulospora (17). In 2014, Marie-Christine Montel et al. studied on cheese products of raw milk in France. They found mold and yeast (18).
In the 2015 survey on milk products in fermented dairy products in Russia, it was shown that the Ascomists, Basidimists, Chytridiomycota and Pichia yeasts are dominant (19).
The same yeasts have been studied in dairy products from around the world in different papers (11,20,24). Based on the variation of milk flora, there is little information of yeasts available compared to bacteria or even aflatoxin producing species. However, the level of health of this fungal survey is very important, especially in cheese-making dairy industry. We used molecular RFLP and sequencing methods in this study. In researches performed on milk, yeasts such as Cand-ida, Rhodotorula, Geotrichum, Kluyveromyces, Terich-osporon, Cryptococcus are usually reported (3). The results we found were consistent with the findings of the study of Karine Lavoie in Canada on 2012. In this study, 67% of the yeasts were identified, and there were many common species (4). Our results were also consistent with the French survey, though not the one cinducted in Canada (18). But it was not similar to the results revealed in the investigation by H. Ksontini et al. in Tunisia (2).
In our study, 15 genera and 31 species were found, most of which were related to Kluyveromyces marxianus and then Pichia kudriavzeii, and then Candida globosa. Note that this is important to us because kluyveromyces is very important for the dairy industry, such as kefir production, and Pichia kudriav-zeii, as a pathogen in human beings. Our results were similar to the 2012 study conducted in Quebec, per-haps due to the common breed of the Holstein. There was a variety of species regarding Candida and C. globosa was the second most prevalent species, but in our research, it was the third most common cause (4).
Also, the flora diversity of each farm was seen unlike the 2006 and 2009 studies that reported the same (25).
Also, our results were similar to those in France. The study, which was conducted in 2011, found Candida, Geotrichum, Cryptococcus, Kluyveromyces, Pichia.-Rodoturola, and Trichosporonspecies. Of course, Geo-trichum, Candida, Kluyveromyces were dominant (26).
We saw more fungal diversity in the Autumn, but it was higher in February in Winter in their study. In their research they also found Yarrowia lipolytica (26).
In many cases, where microflora and milk microbiology have been studied, most of the bacterial species have been noted, and in the case of fungi, the presence of yeast has been reported (2).
Identification of yeasts is important in both pathogenesis and biotechnology. For example, yeasts such as Yarrowia lipolytica is a manufacturer-supplied host for a wide variety of biotechnology applications that are commonly found in some foods and dairy products, such as protein sources for livestock foods, for the production of organic acids or hydrophobic substrates, unsaturated long chain fatty acids and carotenoids (27). But in our study, this yeast was not detected.
Some yeasts such as the ones that can be found in cheese, do not disappear after pasteurization. Since the yeasts C. tropicalis, C. krusei, C. papsilosis, and C. glabrata are pathogenic in humans and animals, and even Trichosporon asahii is pathogenic in immune-compromised individuals, it is important to identify the natural flora of milk. Finally, it is clear that compli-ance with health standards is important for consu-mers. In addition to consumers, it is also important for dairy producers to follow protocols as these same factors can cause mastitis in livestock, which reduces milk supply and increases the cost of keeping and treating livestock.
Conclusion
Understanding the microbial community of the milk helps to identify the environmentally compatible cases as well as its infectious agents, and factors affecting the taste and smell of the dairy products. It also helps to choose more suitable starter to make the flavor more favorable.
Acknowledgements
We would like to thank our colleagues at the Medical Mycology Laboratory, School of Public Health, Tehran University of Medical Sciences, Tehran, Iran.
Conflicts of Interest
The authors reported no conflict of interest.
Received: 2020/12/17 | Accepted: 2021/04/25 | ePublished: 2021/06/28
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |