BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1201-en.html
2- Associate Professor, Razi Vaccine and Serum Research Institute, Agricultural Research, Education and Extension Organization, Tehran, Iran ,
3- Assistant Professor, Department of Microbiology, North Tehran Branch, Islamic Azad University, Tehran, Iran
.
Identifying the bacterial cell-to-cell communication led to an understanding of the fact that interdependent functions are not limited to eukaryotic organisms and can also be present in bacteria. The process of cell-to-cell coordination and communication that involves the production of extracellular and diffuse molecules, followed by the regulation of gene expression, is called quorum sensing (QS) (1). The QS is a process that has changed our view of the life cycle of bacteria, meaning that the bacteria were considered sporadic organisms in the past, and they were believed to ensure their survival through adaptation to environmental conditions without any communication. However, researchers have now discovered the bacterial cell-to-cell communication, signaling and interaction through the information exchange. Therefore, it is concluded that the presence of a signaling network in bacteria is a complex and necessary aspect in their lives (2).
The study of cell-to-cell communication and its effects on the transcription of the unicellular organism has a variety of practical applications. One of them is the possibility of interfering with cell-to-cell communication systems in pathogenic microbes. The QS is a way for bacterial cell-to-cell signaling and exchange information between two partners; the sender and receiver of a communication molecule benefit from the process (3). In bacteria, the QS system regulates phenotypes covering bioluminescence, the production of exoplysaccharides, virulence, conjugative transfer of plasmids, the production of antibiotics and exoenzymes, the formation of biofilms, and the inhibition of growth. Types of molecules involved in communication include acyl homoserine lactones (AHL), autoinducer2 (AI2), and modified oligopeptides (4).
Pseudomonas aeruginosa is an opportunistic and gram-negative human pathogen. These bacteria develop urinary tract infections, respiratory tract infections, skin inflammation and edema, soft tissue infections, bacteremia, bone and joint infections, stomach and intestinal infections, various systemic infections, especially in patients with severe burns, cancer and AIDS having experienced immunosuppression (5). Disruption of the QS system of P. aeruginosa has been shown to eliminate pathogenesis.
Since plants and fungi have had symbiosis with bacteria for millions of years, various mechanisms and substances may have been developed to inhibit the QS system during evolution (6). Garlic is one of the oldest plants whose medicinal effects have long been reported. Garlic has antibacterial effect due to various substances such as alliin, ahoin, allicin and allistain and therefore is consumed against gram-positive and gram-negative bacteria, fungi, parasites and viruses (7).
Since medicinal plants have been used for many years and due to the importance of P. aeruginosa as well as the properties of garlic, the present study aimed to investigate the effect of garlic extract on the QS system of this bacterium in order to use this extract in therapeutic process if there is any inhibition of cell-to-cell communication.
In the present descriptive-analytical cross-sectional study, 12 samples of Pseudomonas were collected from burn wounds of hospitalized patients. The samples were transferred to the laboratory, cultured on the Blood Agar, Eosin Methylene Blue (EMB) and Mueller Hinton Agar (MHA) culture media and then tested morphologically and biochemically.
Identification of Isolates
Colonies were confirmed using standard biochemical and microbiological tests, including gram staining, oxidase, catalase, motility, citrate, TSI, indole, Methyl Red-Voges Proskauer (MR-VP), urease, oxidation-fermentation (OF), growth at 42ºC and pigment formation on Cetrimide Agar medium, and based on the Bergey's manual of determinative bacteriology; all isolates were confirmed as P. aeruginosa (8).
DNA Extraction
The genomic DNA of the isolates was extracted using the GTP kit (Gene Transfer Pioneers, Iran). To this end, 1 to 2 mL of bacterial suspension cultured on EMB equivalent to 0.5 McFarland turbidity standard was used for extraction according to the manufacturer's instructions. The extracted DNA stocks were stored at -20ºC. The quantity and quality of the extracted DNA were examined using spectrophotometer (flow meter E6150) and agarose gel electrophoresis, respectively.
PCR Reaction to Detect the Presence of lasI, lasR and rpoD Genes
The PCR reaction was performed on a final volume of 20 μL containing 3 μm of DNA pattern (200 ng), 10 pmol of each primer (1 μL), 1 μL of distilled water, and 10 μm of 2x Master Mix (Amplicon). The PCR temperature program included the initial denaturation at 95ºC for 3 minutes and the 35 cycle with the denaturation at 95ºC for 30 seconds, the annealing at 55ºC for 1 minute and the extension at 72ºC for 1 minute and the final extension at 72ºC for 10 minutes. Finally, the amplified segments were observed using 1.5% agarose gel electrophoresis and etidium bromide staining. The sequence of primers used is given in Table 1. The PCR reaction was performed using a thermocycler (BioRad-USA).
Table 1. Sequence of primers used in this study
| Primers | Sequence | Length | Reference |
|---|---|---|---|
| lasI-F | GCTTCTGCACGGCAAGGA | 63bp | (9) |
| lasI-R | ATGGCGAAACGGCTGAGTT | 63bp | |
| rpoD-F | GGGCTGTCTCGAATACGTTGA | 90bp | |
| rpoD-R | ACCTGCCGGAGGATATTTCC | 90bp | |
| lasR-F | AAGGAAGTGTTGCAGTGGTG | 68bp | |
| lasR-R | GAGCAGTTGCAGATAACCGA | 68bp |
Preparation of Garlic Extraction
After peeling garlic cloves, 125 g of garlic was crushed using a blender and then mixed with 2 liters of 70% formalin and shaken on the rotator for 48 hours. The resulting mixture was passed through sterile gauze and Whatman filter paper, and the resulting material was centrifuged by a refrigerated centrifuge at 5000 rpm for 30 minutes. Other waste products, such as cellulose and cell shells were removed from the extract and a clear yellowish solution was obtained, distilled for concentration, and stored at 4ºC for subsequent testing.
HPLC Test
In this study, HPLC purification was used to purify the compounds in the immunomodulator fraction of garlic extract and to establish a suitable system for further production of immunomodulators in a shorter time for clinical trials and pharmacological examinations. The fractions collected from the filtration were separated by reversed-phase HPLC on TPV10 208 Vydac C8 reversed-phase preparative column (1 * 25 cm). Purification was performed with 0.1% TFA solution in water and 0.09% TFA solution in acetonitrile and with a flow rate of 1 ml/min from 0% to 15% TFA solutions for 60 minutes. Optical density (OD) was read at the wavelengths of 215, 254 and 280 nm.
Determination of the MIC Value using Broth Dilution Method
The minimum inhibitory concentration (MIC) was determined for each of the bacteria in the exposure to the garlic extract and the tobramycin antibiotic using the standard method proposed by the Clinical and Laboratory Standards Institute (CLSI). The method used a standard 96-well microplate, each containing 100 µL of the Mueller Hinton Broth (MHB) medium. To determine the MIC value, the dilution of garlic extract was started from 2048µg/mL, and 100 µL were added to the first well and then pipetted into the lower wells, and the concentration dropped to the next wells. Next, 100 μL of bacterial suspension equivalent to 0.5 McFarland turbidity standard was poured into all wells. To determine the MIC value for the tobramycin antibiotic, the dilution was started from 32 µg/mL. In all wells, 100 μL of MHB medium was poured and subsequently the first well was added by the antibiotic stock and pipetted to the next wells. After that, 100 μL of microbial suspension equivalent to 0.5 McFarland turbidity standard was added to all wells. In addition, two rows of wells were used as positive control (medium + microbial suspension) and negative control (medium + garlic extract/antibiotic). The microplates were incubated at 37ºC for 24 hours. After this period, the first well with the observed growth was considered as the MIC value (10, 11).
RNA Extraction
In order to investigate the expression of lasI gene using rpoD reference gene, the RNA of Pseudomonas treated with garlic extract and tobramycin antibiotic according to MIC value and also untreated Pseudomonas was extracted using Cinna Pure-RNA kit (CinnaGen, Cat. No. PR891620, Iran). Quantus™ Fluorometer (Cat.# E6150) and Promega kit were used to quantify the RNA extracted.
Construction of cDNA
To this end, 1 µg of RNA was mixed with 1 µL of enzyme buffer, 1 µL of DNase enzyme and 0.5µl of RNase inhibitor, respectively, and then reaching final volume of 10 µL with RNase-free water. The microtubes were incubated at 37°C for 30 min and 1 µL of EDTA 0.5M was added to each. The incubation was performed for 10 min at 65°C to inactivate the DNase enzyme; 0.5 µL of random hexamer and 0.5µL of Oligo (dT) were added to the microtubes and incubated at 65°C for 5 min. After this time, the microtubes were transferred onto the ice and 13 µL of this MasterMix was added to each. The microtubes were incubated at 25°C for 5 min, at 42°C for 60 min, and at 70°C for 10 min. The resulting single-stranded cDNA was stored at -20ºC. The used MasterMix contained 1 µL of dNTP, 4 µL of RT buffer, 2 µL of RT enzyme, 0.5 µL of RNase inhibitor and 5.5 µL of RNase-free water.
Real-time PCR Process
To perform the real-time PCR process, 10 µL of PCR MasterMix, 1 µL of each of the forward and reverse primers (Table 1), 1.5 µL of constructed cDNA and 5.6 µL of water were mixed in a microtube. Samples were placed in the device with a temperature program including 10 minutes of initial denaturation at 94ºC, 15 seconds at 94ºC, 25 seconds at 55ºC and 35 seconds at 72ºC.
Real-time PCR Data Analysis
The real-time PCR data were analyzed by the software available in the one plusABI device using the –∆∆CT method, and the final number (2–∆∆CT) obtained from various repeats was statistically analyzed using the GenEX software
HPLC test
Evaluation of garlic extract using HPLC method and analysis of the resulting chromatogram showed that the highest compound in the extract is allicin. This study was compared with the internal standard (Figure 1).

Figure 1- HPLC chromatogram for allicin and internal standard (IS 2.7.4. Polymeric nanoparticles
MIC Determination
After 24 hours of incubation at 37ºC, the sub-MIC value was determined to be 256µg/mL for garlic extract and 16µg/mL for tobramycin.
Investigating the Presence of lasI, lasR and rpoD Genes
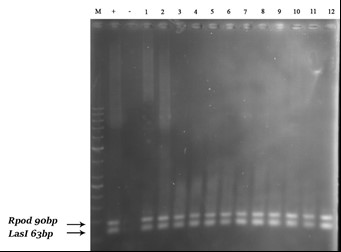
The results of PCR molecular analysis for the lasI, lasR and rpoD genes of QS system were obtained on the basis of band observation in the mentioned region on 1% agarose gel. In the present study, all 12 (100%) samples of P. aeruginosa isolated from clinical samples had these genes (Figures 2 and 3).
Figure 2. PCR test results for lasI and rpoD genes on a number of strains; left to right, respectively: 100-bp DNA marker plus, positive (+) control, negative (-) control, samples
Figure 3- PCR test results for lasR gene on a number of strains; left to right, respectively: 100-bp DNA marker pluse, positive (+) control, negative (-) control, samples
Analysis of Gene Expression
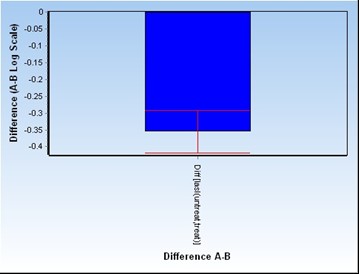
The real-time PCR technique was used to determine the rate of suppression of gene expression quantitatively in the presence of garlic extract and tobramycin. The real-time PCR process was performed for the lasI gene using the rpoD reference gene, the results of which are shown in Figures 4 (in the presence of garlic extract) and 5 (in the presence of tobramycin). The results indicated a significant difference in lasI gene expression between the two groups untreated and treated with garlic extract. The fold change rate for the lasI gene was calculated to be -1.19, indicating that this gene was reduced up to 1.19 times in the group treated with garlic extract compared to the untreated group. The expression of lasI gene in the tobramycin-treated group was also significantly reduced. The fold change rate for the lasI gene was calculated to be -1.27, indicating that the gene in the tobramycin-treated group decreased up to 1.27 times compared to the untreated group. There was no significant difference between the effects of garlic extract and tobramycin in terms of their ability to reduce lasI gene expression.
Figure 4. Diagram of -∆∆CT in the presence of garlic extract
Figure 5. Diagram of -∆∆CT in the presence of tobramycin antibiotic
The spread of bacterial infections and the development of their antibiotic resistance have prompted researchers to find natural antibacterial compounds. These studies are often performed on various plant compounds, their extracts, and their active compounds (12). The aim of this study was to investigate the effect of garlic extract on P. aeruginosa bacteria by evaluating the effect of garlic extract on the expression level of genes affecting bacterial QS system and in comparison with tobramycin antibiotic.
Some garlic compounds include alliin, allicin, organic acids, carbohydrates and vitamins; and the most important properties of garlic are related to allicin (13). The allicin is an essential oil with a bright yellow color and responsible for the specific aroma of garlic. The allicin is also called allicin potential or allicin yield and accounts for the antimicrobial properties of garlic. This compound plays a major part in its antibiotic properties through specific inhibition of acetylcholine A synthase; the inhibition of this enzyme inhibits lipid biosynthesis and fatty acids and ultimately disrupts the bioavailability of microorganisms (14). One of the important features of allicin is its ability to penetrate and pass through membrane phospholipids, which is why it can freely pass through the membrane and perform its functions (15). In the analysis of garlic extract obtained in the present study using HPLC technique, the allicin was the main ingredient in the extract. Therefore, its antimicrobial effects were expected to be observed on P. aeruginosa, but the main objective was to evaluate the mechanism of action of this antimicrobial agent and to investigate the expression of QS-related genes and the biofilm formation in the presence of extract.
The lasI and lasR genes are inherently present in the genome structure of most pathogenic P. aeruginosa bacteria isolated from medical centers and develop QS system, biofilm and drug resistance in these strains. In the P. aeruginosa bacteria, the QS system consists of two gene systems, LasR-LasI and RhlR-RhlI. The LasI and RhlI genes express acyl-hemoserine lactone (acyl-HSL) synthase, while the LasR and RhlR genes produce transcriptional regulatory proteins, which bind to their specific signals and activate target (virulence) genes (16). The QS system also exists in other species of Pseudomonas, but the number and sequence of their structural genes varies. Many genes in P. aeruginosa are controlled and expressed by QS, which are involved in pathogenesis, including lasI, lasR, rhlI, rhlR genes, and controlled genes called lasB, apr, and rhlAB in clinical isolates (17).
Plant compounds have been shown to inhibit biofilm formation in a variety of ways. Herbal extracts with bacteriostatic or bactericidal properties inhibit or reduce bacterial biofilm formation. However, some herbal compounds affect biofilm formation without bacteriostatic or bactericidal effects. Their advantage is the failure of bacteria to resist these compounds. For example, some plant compounds interfere with the QS system, which controls bacterial biofilm (18).
The garlic extract stops the QS system and accelerates the elimination of lung infection caused by P. aeruginosa. The garlic extract was used to treat lung infections in rats and it was found that the garlic therapy first stimulated higher levels of inflammation and significantly eliminated the pathogenic bacteria (19). Fulghesu et al. (2007) examined different QS inhibitory compounds in P. aeruginosa. They tested the inhibitory activity of three macrolide drugs and three lincosamide drugs, resveratrol, garlic extract and N-acetyl cysteine on four strains of P. aeruginosa isolated from patients with cystic fibrosis. All of the compounds tested were able to inhibit QS in the tested strains; only lincomycin and N-acetyl cysteine did not inhibit QS in one of the Pseudomonas strains (20).
In another study, the QS inhibition was investigated using a new genetic system. A set of QS inhibitor screening systems was developed that enabled them to identify a number of new natural and synthetic inhibitors. The two active inhibitors identified in this study were garlic extract and 4-nitropyridine-N-oxide (4-NPO). Transcriptome analysis based on gene arrays showed that 4-NPO and garlic extract were specific for QS-controlling virulence genes in P. aeruginosa. The two QS inhibitors also significantly reduced the biofilm resistance of P. aeruginosa compared to tobramycin (21). The QS inhibition activity was investigated by garlic extract and QS response reduction was observed in QS receptors of Escherichia coli, Agrobacterium tumefaciens, Chromobacterium violaceum, Pseudomonas putida and Pseudomonas chlororaphis in relation to growth inhibitory effects (22). In addition, the QS inhibition and pathogenesis attenuation of P. aeruginosa were investigated under the influence of garlic extract and it has been shown that oral treatment with garlic extract significantly reduces the density of pulmonary bacteria. In vitro experiments also showed that the virulence factors and the formation of QS signals by the bacterium P. aeruginosa decreased in the presence of fresh garlic extract (23).
Accordingly, the effect of garlic extract on the QS system of P. aeruginosa has been proven frequently, but none of the studies mentioned have expressed the mechanism of this effect and examined the expression of QS genes. In the present study, the expression level of lasI gene as one of the important genes in bacterial QS system was determined by real-time PCR technique, the results of which showed that the garlic extract at a concentration of 256 μg/mL (as MIC value) decreases the expression of lasI gene. In order to compare decreasing effect of garlic extract on the expression of these genes with the effect of tobramycin, the expression of lasI gene in the presence of this antibiotic was also investigated, which showed a greater decrease compared to the garlic extract, but not significant. Therefore, the promising results are that the active ingredients of garlic can be extracted and used as adjunctive antibiotic therapy or as an independent treatment in infections. This active ingredient was found to be allicin in the present study. It is recommended that the extracted and purified allicin in future research should be applied as an anti-QS and therefore antibacterial agent for the treatment of Pseudomonas aeruginosa infections.
The authors declared no conflict of interests
Received: 2020/08/9 | Accepted: 2020/10/13 | ePublished: 2021/01/10
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |