BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1052-en.html
2- Department of Biology, Parand Branch, Islamic Azad University, Parand, Iran ,
3- Department of Chemical and Petrochemical Engineering, Sharif University of Technology, Tehran, Iran
Staphylococcus aureus is a pathogenic bacterium of the Micrococcus family, which is a common pathogenic bacterium in hospital infections (1, 2). It causes a wide range of diseases including skin infections, pneumonia and endocarditis (3). One of the opportunistic pathogenic agents of nosocomial infections is methicillin-resistant Staphylococcus aureus (MRSA). Resistance to antibacterial agents in MRSA strains has caused limitations in the treatment of diseases caused by this bacterium (4).
One of the most important mechanisms of antibiotic resistance in bacterial cells is the outflow pumps that cause antibiotic outflow from bacterial cells, which is observed in many clinical pathogens (8). Output carriers of antibiotics are divided into five main families based on the amino acid sequence. These families include the major facilitator superfamily (MFS), the Resistance-nodulation-division (RND), the small multidrug resistance (SMR), the ATP-binding cassette enzyme-linked family, and multiple antibiotics, and the extrusion antibiotic family (9).
One of the important mechanisms of antibiotic resistance in S. aureus is the presence of efflux pumps. NorA, norB and norC efflux pumps are found in drug-resistant strains (19). Various studies have shown that norA is capable of pumping various compounds including hydrophobic fluoroquinolones such as norfloxacin, ciprofloxacin and ethidium bromide (22). There is a direct relationship between increased expression of norA efflux pump and increased resistance to fluoroquinolones. Inhibition of these systems can be a promising strategy to enhance the effectiveness of antibiotics (14).
Many studies have shown that a wide range of plant extracts may act against bacterial resistant mechanisms (15). Artemisia belongs to the family asteraceae, a small plant that grows in temperate regions. It has 34 species in Iran and is highly regarded in Iranian and Chinese traditional medicine. It has numerous biological properties including antibacterial, antifungal, antioxidant and anti-inflammatory (16).
The aim of this study was to investigate the effect of A. ciniformis extract on expression Efflux pump norA in clinical isolates of ciprofloxacin-resistant S. aureus, a prospective study in Iran which was not studied before.
This study was done using clinical samples collected in authors’ previous study (18). Collecting clinical specimens, detecting ciprofloxacin-resistant S. aureus isolates, performing microbial susceptibility testing (antibiogram) by agar disk diffusion method, PCR detection of norA gene, and phenotypic evaluation of active efflux pump was as presented in previous study (18).
Collecting and Extracting A. ciniformis
The A. ciniformis was prepared from the Iranian Biological Resource Center and kept in optimum condition. To do the extraction, 40 g of the plant was first weighed and 300 mL of 80% ethanol was added to the plant and kept in a shaker system for 24 h (90 rpm). After 24 h, it was filtered and placed in an incubator at 37°C to completely evaporate the alcohol.
Determination of Minimum Inhibitory Concentration (MIC) of Plant Extract
MIC experiments were performed according to the dilution method in the microplate for the extract. MIC was performed three times by microdilution method on 96 well plates. The extract was poured into wells at concentrations of 15.6 to 1000 µg / mL and with Mueller Hinton Broth (MHB) it reached to the concentration of 195 µL. All wells were added with 5 µL of microbial culture of isolates at half McFarland concentration. The MIC value was considered as the lowest inhibitory concentration of bacterial growth. It should be noted that wells containing non-extracted bacteria were considered as negative controls and wells containing standard bacteria S. aureus ATCC 25923 and extracts were considered as positive controls to determine MIC concentration.
Extraction of RNA from Staphylococcus aureus Treated with subMIC Extract
Bacterial cell lysis and RNA extraction were performed using RNX solution (Sinagen) according to the protocol. The cDNA construct was performed according to the Revert AidTM First Strand cDNA Synthesis Kit (Fermentas). The gmk gene (guanylate kinase) was used as the reference gene. The primers of the gmk and norA genes are presented in Table 1.
Real-time PCR reaction in the final volume of 25 µL was optimized as follows: 12.5 µL of SYBR TM (2X) Master Mix (Takara Company), 1 µL of 5 pmol primer (Takapoo Biol.), 9.5 µL of deionized water and 1 µL cDNA template (100 ng). Thermal program was performed as follows with Bioneer exicycler 96: Primary denaturation of DNA template was performed at 95°C for 10 min, and the second step alternately was performed during 40 cycles at 95°C for 20 seconds and at 61°C for 40 seconds and 72°C for 20 seconds.
Data Analysis
Statistical calculations were performed using Graphpad Prims ver.6 software and the results were analyzed by one way ANOVA. The difference of target genes expression between control and treated samples was calculated by Tukey's HSD post-hoc test. Data were presented as mean ± standard deviation (SD) and P<0.05 was considered significant. Real time PCR data analysis was performed based on the comparison of the threshold cycle with the following formula:
ΔCt= Ct target-Ct reference
ΔΔCt=ΔCt test sample - ΔCt control sample
Relative expression: 2 –ΔΔCt
Table 1. Primers used in this study
| PCR products (bp) | sequences | primers |
| 112 | 5'ATCGGTTTAGTAATACCAGTCTTGC3' | norA-F |
| 5'GCGATATAATCATTTGAGATAACGC3' | norA-R | |
| 188 | 5'TATCAGGACCATCTGGAGTAGG3' | gmk-F |
| CATCAACTTCACCTTCACGC | gmk-R |
MIC Results of Artemisia ciniformis Extract Against Ciprofloxacin-resistant Strains
According to the findings of the previous study, out of 250 clinical specimens, 50 isolates of Staphylococcus aureus were isolated and antibiotic susceptibility results showed that 68% of the samples (34 samples) were methicillin resistant and 24% of them (12 samples) were ciprofloxacin resistant and none of the samples were vancomycin resistant (18).
Ciprofloxacin-resistant S. aureus positive isolates were affected by concentrations of 15-1.6.6 µg / mL of the plant extract over a 24-hour period. The results showed that different strains had a range of MIC of 31.55-500 µg / mL (Table 2).
The graphs of the amplification of norA and gmk genes are shown in Figures 1 and 2. Specific amplification of the target gene fragments, primers not pairing, and no amplification of non-specific fragment for each gene were determined using the melting curve (1-2)The results showed that the expression of norA gene was significantly decreased compared to the reference gene (gmk) after treatment with the plant extract. The highest decrease in norA gene expression was observed in isolate 6 and the lowest decrease in gene expression was observed in isolate 5. The results are shown in Figure 3.
Table 2. Determination of MIC of plant extract in different strains.
| Isolate number | MIC (µg/mL) |
| 1 | 62.5 |
| 2 | 125 |
| 3 | 31.2 |
| 4 | 250 |
| 5 | 500 |
| 6 | 15.6 |
| 7 | 62.5 |
| 8 | 31.2 |
| 9 | 125 |
| 10 | 500 |
| 11 | 31.25 |
| 12 | 250 |
| ATCC 35556 | 500 |
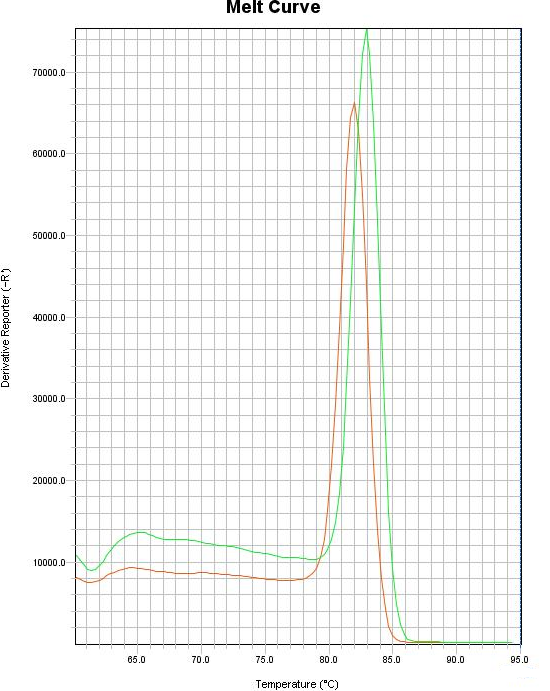
Figure 1. Diagram of melting curves of norA genes and gmk genes.
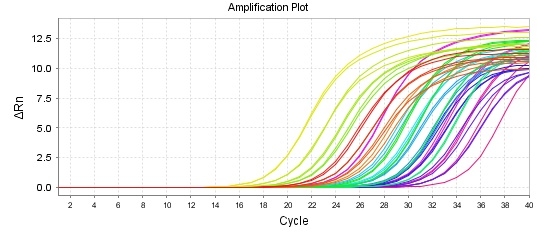
Figure 2. Amplification plot of norA and gmk gene in bacterial cells treated with extract.

Figure 3. NorA gene expression in extract-treated strains.
Since ancient times, medicinal plants have been used clinically as important sources of secondary metabolites (terpenoids, phenolic compounds and alkaloids). These plants can directly or indirectly affect the activity of efflux pumps. Recently, the use of herbal compounds and plant extracts as an alternative to antibiotics has received much attention (26). The difference in the effect of plant extracts on bacteria depends on various factors. In general, plant compounds exert their antimicrobial activity through a variety of mechanisms (27).
Due to their antibacterial potential, the use of herbal extracts can be an effective, low cost and affordable method to reduce antibiotic resistance in a wide range of hospital opportunistic bacteria (28).
The results of our study showed that A. ciniformis extract was able to significantly reduce norA gene expression in ciprofloxacin resistant strains. It should be noted that different expression of norA gene was observed in the resistant strains and in the more resistant strains a higher relative expression of the norA gene was observed.
Flavinoid sarothrin extracted from Alkann orientalis leaves and flowers inhibited norA efflux pump activity and inhibited the growth of Mycobacterium smegmatis and S. aureus (28).
The results of the study by Kalia et al. showed that capsaicin decreased the MIC of ciprofloxacin for S. aureus SA-1199B (norA overproducing), SA-1199 (wild-type). Increased susceptibility of S. aureus to ciprofloxacin was observed. This study showed that capsaicin not only decreased the efflux pump activity but also decreased the attack of Staphylococcus on macrophages (29).
Studies have shown that the extract and compounds of Olympicin A, 5,7-dihydroxy-6- (2-methylbutanoyl) -8- (3-methylbut-2-enyl) -4-phenyl-2H-chromen-2-one, Karavilagenin C and Ailanthoidiol, Boeravinone B from aerial parts of Hypericum olympicum L., flowers of Mesua ferrea L., aerial parts of Momordica balsamina L., root of Zanthoxylum capense and Boerhavia diffusa inhibited and decreased activity of norA efflux pump. In other words, they had an Efflux pump inhibitor (EPI) (30-34).
This study was in line with other studies showing that A. ciniformis reduced norA gene expression in efflux pumps in S. aureus. Decreased norA gene expression leads to poor performance of these pumps and prevents overflow of antibiotics and other disinfectants and pharmaceuticals. Therefore, this plant can be used as an appropriate antibacterial drug and, on the other hand, it is recommended with regard to its antiviral resistance, other efflux studies and antiviral antibiotic resistance.
The authors would like to acknowledge the laboratory of Islamic Azad University.
Authors declared no conflict of interests.
Received: 2019/10/2 | Accepted: 2020/01/17 | ePublished: 2020/03/14
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |







