BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-953-en.html
2- Department of Food Microbiology, Faculty of Food Sciences and Industries, Gorgan University of Agricultural Sciences and Natural Resources, Gorgan, Iran ,
.
The term “foodborne diseases” is more commonly referred to as food poisoning, is used to describe the gastrointestinal side effects that occur following the consumption of certain foods or drinks. Foodborne illnesses affect 48 million people in the United States each year (1-3). There are more than 200 identified diseases that can be transmitted through food and by various factors such as bacteria, fungi, viruses, and parasites. According to food safety and health experts, millions of people around the world each year are affected by foodborne pathogens. While food supply and production in the United States is one of the safest in the world, the US Centers for Disease Control and Prevention estimate that foodborne illnesses cause 76 million illnesses, more than 300,000 per year hospitalization, and 5,000 deaths in the United States (4-6). This figure is also estimated to be 2366,000 in England and Wales, 21138 hospitalizations, 718 deaths (7). On the other hand, with the frequent spread of diseases caused by new pathogens, the use of antibiotics in livestock breeding and resistance gene transfer to human, and current concerns about bovine spongiform encephalitis are just a few examples of these risks (8).
Thus, despite extensive advances in health and food safety food borne pathogens still cause many poisoning and digestive problems for consumers. Listeria monocytogenes, Staphylococcus aureus, Salmonella enterica and Escherichia coli are food borne pathogens that cause problems for consumers for various reasons such as lack of hygiene before, during or after food production.
Over the past decade, there has been a growing interest in improving the quality and enhancement of food safety by replacing conventional conservation and maintenance systems with natural alternatives. Bio-preservatives are defined as "the use of microorganisms or their metabolites to prevent spoilage, enhance food safety and shelf life" (9,10). Lactic Acid Bacteria (LAB) are bio-preservatives that play a key role in the fermentation process (11). In addition, it has been shown that LAB have antimicrobial activity in fermented foods and therefore can be used as natural preservatives to inhibit the growth of food born pathogenic bacteria and fungi (12,13). Lactic acid bacteria produce a variety of metabolites including organic acids, bacteriocins, hydrogen peroxide and low molecular weight metabolites such as diacetyl and reuterin with inhibitory effects on Gram-negative and Gram-positive producing food spoilage bacteria (such as Micrococcus, Pseudomonas, Moraxella, Acinetobacter, Shewanella( and as well as food poisoning bacteria (such as S. aureus, L. monocytogenes, Clostridium botulinum type E, Yersinia enterocolitica) (14). The mechanism of action of bacteriocins is to destabilize the cells and increase the permeability of the cytoplasmic membrane. Lactic bacteria produce a wide range of bacteriocins (nisin, pediocin, lactacin, divergine, diplocin, elactosterone) that have a different antimicrobial spectrum but are more likely to be effective on Gram-positive pathogenic and food spoilage bacteria (15). In this study, we tried to evaluate the antimicrobial potential of a number of lactic acid bacteria isolated from traditional products against some food-borne pathogens.
Bacterial Strains
In this study, bacteria isolated from local yogurt and milk and camel dooq (yogurt drink) of Golestan province with codes M109, M8, Y92, Y91, 8C, Y102, M153, Y73, Y52, Y98, Y89 that were prepared from microbial collection in Department of Food Science and Technology of Gorgan University, were used. To investigate the antimicrobial properties of the pathogenic bacteria, L. monocytogenes (ATCC 19115, PTCC 1298), S. enterica (ATCC 14028, PTCC 1709), E. coli (ATCC 25922, PTCC 1399) and S. aureus (ATCC 25923, PTCC 1431) were used. All pathogenic bacteria were purchased from the Persian Type Culture Collection (PTCC), Tehran, Iran.
Activation of Isolates and Phenotypic Identification
For this purpose, the isolates were first cultured in MRS broth under anaerobic conditions at 37°C. Identification of the isolates in early stages was performed using phenotypic criteria such as colony shape and cell morphology, Gram staining and catalase activity. MRS agar and MRS broth and yeast extracts were prepared from Merck, Germany and Muller Hinton Agar (MHA) from Sigma, USA.
Molecular Identification of Isolates
The DNA extraction was carried out using a commercial DNA extraction kit (Takapouzist, South Korea). To identify molecularly LAB by PCR, Universal primers (1492R: 5´-GGTTACCTTGTTACGACTT-3´ and 27F: 5´-AGAGTTTGATCCTGGCTCAG-3´) were used to amplify 16S rDNA variable regions. The length of the amplified DNA fragment was 1500 bp.
The PCR was performed in a volume of 30 µL with optimized amounts of 15 µL of Red 2X Master Mix (Macrogen, South Korea), 5.5 µL of each primer, 3 µL of DNA and 11 µL of water in a thermo cycler (Corbett research, CG1-96, Australia).
PCR products were sent to Macrogen Company in South Korea for sequencing. Sequences were compared with available sequences in the world gene bank (NCBI) using the Blast program. Isolates with the highest percentage of similarity of their sequences with the stored sequences in the gene bank were identified as the same species.
Evaluation of Antibacterial Activity of the Isolates
The micro-dilution method was used to evaluate the antimicrobial activity of the Cell Free Spent Medium (CFSM) of 24-hour culture of lactic isolates against pathogenic bacteria including L. monocytogenes, S. aureus, S. enterica and E. coli. At first, LAB were cultured in MRS broth under anaerobic conditions at 37°C. At the end of the logarithmic phase, CFSMs were centrifuged at 6,000 rpm for 10 min and then filtered with a 0.45 μm syringe filter and were used for antimicrobial testing. To determine the antimicrobial effect, 185 μL of CFSM of each lactic isolate along with 15 μL of pathogenic bacteria (105 cfu/mL) were added into each well. After 24 h of incubation under aerobic conditions at 37°C, the samples absorbance were measured at 600 nm. The inhibitory percentage of lactic isolates was calculated as follows:
1- [the growth in test well/ the growth in positive control well] × 100
Statistical Analysis
The results of this study were analyzed by one-way ANOVA and LSD at the significant level of 0.05 in three replications using SPSS 19 (SPSS In., Chicago, IL., USA) and Microsoft Excel 2007 software was used to draw charts
Molecular Identification of Activated Isolates
To confirm DNA replication, the PCR products were loaded onto gel electrophoresis (Figure 1).
As shown in Figure 1, the PCR products of each of the 11 extracted DNA samples had a length of 1500 bp. After blasting the PCR product sequences with the data in the NCBI database, it was found that the isolates were Lactobacillus acidophilus, Lactobacillus fermentum, Enterococcus faecium, Lactobacillus brevis and Lactobacillus rhamnosus (Table 1). Based on the above results and the obvious differences between molecular and biochemical identification, it can be concluded that molecular identification of microbial strains using the 16S rDNA gene region is the most accurate method.

Figure 1. Gel Electrophoresis of PCR products containing specific primer with 500 Bp target sequence on 1.5% gel agarose. 1: Marker, 2: Positive control (Lactobacillus plantarum), No. 12 to 22: activated samples from 1 to 11, respectively.
Table 1. Sequencing results of activated isolates and comparison of results obtained from biochemical and molecular identification
| No. | Isolate code | source | Molecular identification | Biochemical identification |
| 1 | Y92 | Local yogurt | Lactobacillus brevis | Lactobacillus spp. |
| 2 | M153 | Local milk | Lactobacillus rhamnosus | Lactobacillus spp. |
| 3 | C8 | Camel dooq | Enterococcus faecium | Pediococcus pentosaceus |
| 4 | M109 | Local milk | Enterococcus faecium | Enterococcus spp. |
| 5 | Y98 | Local yogurt | Lactobacillus acidophilus | Lactobacillus spp. |
| 6 | Y52 | Local yogurt | Lactobacillus brevis | Lactobacillus spp. |
| 7 | M8 | Local milk | Enterococcus faecium | Enterococcus spp. |
| 8 | Y91 | Local yogurt | Lactobacillus reuteri | Lactobacillus spp. |
| 9 | Y102 | Local yogurt | Lctobacillus rhamnosus | Lactobacillus spp. |
| 10 | Y73 | Local yogurt | Enterococcus faecium | Enterococcus hirae |
| 11 | Y89 | Local yogurt | Lactobacillus rhamnosus | Lactobacillus spp. |
Antibacterial Activity of the Isolates
Among the identified isolates, 11 isolates of different genera and species were selected and then their antimicrobial activity against pathogenic bacteria was investigated by micro-dilution method (Figures 2 to 5). The results of the evaluation of the inhibitory effects of the CFSM of lactic isolates on the growth of L. monocytogenes (Figure 2) showed that the percentage of inhibition of the CFSM of the lactic isolates were varied from 29.96 to 30.99 percent. Among the isolates, E. faecium C8 isolated from camel dooq showed the highest inhibitory percentage (P<0.05). Also, no significant difference was observed in the inhibitory percentage among L. rhamnosus Y89, E. faecium M109 and L. reuteri Y91. Also, L. brevis Y92 had the lowest inhibitory percentage (P<0.05).
The results of the evaluation of the inhibitory effects of CFSM of lactic isolates on the growth of S. aureus are shown in Figure 3. As shown, L. rhamnosus Y89 and E. faecium M109 had the highest percentage of inhibition against S. aureus. Also E. faecium Y73 had significantly the least inhibitory effect (P<0.05).
In another part of this study, the inhibitory effects of the CFSM of lactic isolates on the growth of two gram-negative bacteria including E. coli (Figure 4) and S. enterica were investigated (Figure 5). The results showed that the inhibitory rate of the isolates on the growth of E. coli and S. enterica varied from 2.42 to 36.93 percent and from 1.14 to 31.97 percent, respectively. Also, the highest inhibitory effect on E. coli belonged to E. faecium M109 isolated from milk and the lowest inhibitory rate belonged to L. brevis Y92 isolated from yogurt (P<0.05).
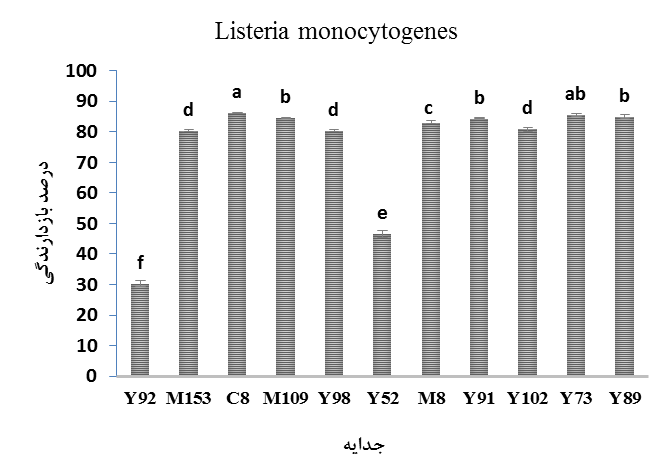
Figure 2. Inhibitory effects of CFSM of lactic isolates on growth of Listeria monocytogenes
* Similar letters in each column indicate no significant difference at 0.05 level.
* Y: Local yogurt; M: Local milk; C: Camel dooq

Figure 3. The inhibitory effects of the CFSM of lactic isolates on the growth of Staphylococcus aureus
* Similar letters in each column indicate no significant difference at 0.05 level.
* Y: Local yogurt; M: Local milk; C: Camel dooq
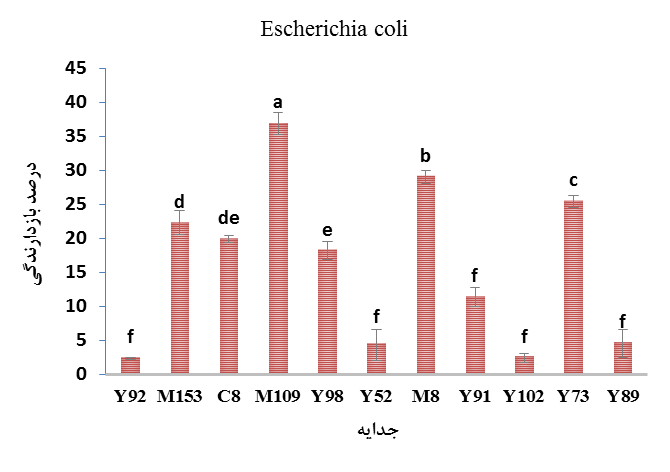
Figure 4. The inhibitory effects of the CFSM of lactic isolates on the growth of Escherichia coli
* Similar letters in each column indicate no significant difference at 0.05 level.
* Y: Local yogurt; M: Local milk; C: Camel dooq

Figure 5. The inhibitory effects of the CFSM of lactic isolates on the growth of Salmonella enterica
* Similar letters in each column indicate no significant difference at 0.05 level.
* Y: Local yogurt; M: Local milk; C: Camel dooq
Table 2. Comparison of inhibitory effect of lactic isolates on pathogenic bacteria
Similar letters in each row indicate no significant difference at the 0.05 level.
| Salmonella enterica | Escherichia coli | Staphylococcus aureus | Listeria monocytogenes | Isolate code | No. |
| 0.64b±1.4 | 0.15b±2.44 | 0.17 a±28.12 | 0.92 a±30.29 | Y92 | 1 |
| 0.9 d±18.46 | 1.76c±22.29 | 0.69 b±71.17 | 0.42 a±80.46 | M153 | 2 |
| 0.74 c±20.93 | 0.53c ±19.96 | 0.43 b±59.11 | 0.085 a±86.14 | C8 | 3 |
| 1.21 d±31.97 | 1.61c±36.93 | 1.04 b±81.59 | 0.08 a±84.48 | M109 | 4 |
| 0.55 c±16.74 | 1.36c±18.21 | 0.6 b±71.71 | 0.42 a±80.46 | Y98 | 5 |
| 0.37 c±4.31 | 2.29c±4.41 | 3.12 b±33.15 | 1.25 a±46.56 | Y52 | 6 |
| 1.32 c±30.34 | 0.95c±29.08 | 1.48 b±77.67 | 0.5 a±83.06 | M8 | 7 |
| 0.52 c±13.67 | 1.4c±11.44 | 1.3 b±68.14 | 0.17 a±84.4 | Y91 | 8 |
| 0.68 c±3.78 | 0.67c±2.5 | 0.26 b±59.98 | 0.67 a±80.87 | Y102 | 9 |
| 0.83 d±13.95 | 0.88b±25.43 | 0.13c±21.38 | 0.59 a±85.49 | Y73 | 10 |
| 0.46b±5.65 | 2.07 b±4.63 | 0.17 a±84.71 | 0.67 a±84.9 | Y89 | 11 |
According to the results of this study, Lactobacillus and Enterococcus genera were isolated and identified as lactic isolates from traditional fermented dairy products. Other studies have also confirmed the presence of these genera in traditional and native dairy products. In this regard, research by Haghshenas et al. (2014) indicated that they were able to identify Enterococcus mundtii 50H, Enterococcus daurans 39C and Enterococcus faecalis 13C in traditional dairy products (18). Tulumoğlu et al. (2014) also identified 7 strains of Lactobacillus fermentum in a study of more than 100 isolates from Tulum cheese (19). Research by Leite et al. (2015) on the study of lactic acid bacteria from Brazilian kefir grains led to the identification of Leuconostoc mesenteroides, Lactococcus lactis, Lactobacillus paracasei isolate and Lactococcus lactis subsp. lactis (16). Thus, the Lactobacillus and Enterococcus genera are the most lactic acid bacteria isolated from traditional and native dairy products. The results of evaluation of the inhibitory effect of lactic isolates on L. monocytogenes and Staphylococcus aureus showed that all isolates had good antibacterial activity against these two pathogens and the highest inhibition percentage was related to C8 isolate (isolated from camel dough) and Y89 (isolated from local yogurt), respectively. These results are consistent with the findings of Leite et al. (2015) who showed that the lactic isolates of Lactococcus lactis, Leuconostoc mesenteroides and Lactobacillus paracasei have inhibitory effect on L. monocytogenes, S. aureus, S. enterica and E. coli (16). The inhibitory properties of lactic isolates can be attributed to various antimicrobial metabolites such as lactic acid, acetic acid, hydrogen peroxide, carbon dioxide and bacteriocin (20, 21). Casaburi et al. (2016) also isolated Lactobacillus curvatus 54M16 from Campania's traditional fermented sausage. It produced more than one bacteriocin, including saccharin X, T and P, and showed inhibitory effects on L. monocytogenes, Bacillus cereus, and Brochothrix thermosphacta (22). The results of the inhibitory effects of lactic isolates on the growth of E. coli and S. enterica also showed that all strains identified from traditional Iranian dairy products except a few strains were able to inhibit the growth of mentioned pathogenic bacteria and the highest inhibition percentage was related to M109 isolate (isolated from local yogurt). In addition, it was found that, among all the isolates, the highest inhibition percentage was related to C8 isolated from camel dough. Also, the cause of less resistance of Gram-positive bacteria than Gram-negative bacteria can be attributed to the impermeable wall and the complex and multilayer structure of these bacteria as well as the presence of an outer membrane. Sabir et al. (2010) also isolated L. acidophilus strain Z1L from kefir, which was able to significantly inhibit E. coli growth (23). Paris Silia et al. (2015) also evaluated the antimicrobial effect of different species of Lactobacillus genus from traditional Cucido Mexican cheese. Their results showed that the bacteriocin-like compounds produced by these bacteria exhibit significant antimicrobial activity against Listeria innocua, Staphylococcus aureus, Salmonella typhimurium, and Escherichia coli. Recently, numerous reports have been published on the antimicrobial activity of lactic acid isolates (24-26). For example, Angmo et al. (2016) isolated and identified Lactobacillus plantarum KJ722784 from Ladakh and Assohoun et al. (2016) isolated and identified Lactobacillus fermentum from Doklu that both isolates were able to produce antimicrobial compounds including bacteriocins (24, 25).
Bacteriocins inhibit cell wall synthesis by binding to lipid II, leading to cell death by removing lipid II from the membrane structure and pore formation. They also inhibit DNA replication by blocking the activity of DNA gyrase enzyme, destroying DNA by nuclease activity, and blocking protein synthesis by disrupting ribosomal activity (27). According to the results of Schillinger et al. (2005), L. acidophilus had significant accumulation potential against pathogenic bacteria due to its high level of hydrophobicity on the cell surface. There is a direct relationship between the rate of cell wall hydrophobicity and the ability of lactic acid bacteria to accumulate against pathogenic bacteria (28). L. acidophilus and Lactobacillus gasseri isolated by Fernandez et al. (2003) were also able to prevent the growth of Salmonella, Listeria and Campylobacter bacteria without interfering with the microbial flora of the gastrointestinal tract (29). Lactic acid bacteria also have the ability to decrease cholesterol levels, immune system immunization and anti-tumor activity (30).
Due to the importance of specific properties of lactic isolates from traditional and indigenous dairy products, identifying and evaluating the potential properties of these isolates is of great importance.
According to the results of this study, lactic strains isolated from traditional dairy products can be used separately or in combination with other preservatives to reduce the consumption of synthetic preservatives or as starter culture in the food industry.
The sequencing results of PCR products led to the identification of Lactobacillus and Enterococcus genera from traditional fermented dairy products. Also the results of evaluation of the inhibitory effect of Lactic isolates on the growth of L. monocytogenes and S. aureus showed that all isolates were able to prevent the growth of these two pathogens and their inhibitory percentages varied from 86.14 to 14.14 percent. L. rhamnosus Y89 isolated from yogurt and E. faecium isolated from camel dough showed the highest inhibition percentage on L. monocytogenes and S. aureus, respectively.
Comparison of the inhibitory effect of lactic isolates on pathogenic bacteria showed that the inhibitory effect of all tested lactic isolates on Gram-positive bacteria was significantly (P<0.05) more than their effect on Gram negative bacteria. According to the results of this study, all strains isolated and identified from these dairy products are able to prevent the growth of Gram-positive pathogenic L. monocytogenes and S. aureus and except for some strains, all lactic isolates have the ability to inhibit the growth of Gram-negative bacteria E. coli and S. enterica. Therefore, it is suggested that the lactic isolates obtained from yogurt, milk and camel dough be used as bio-preservatives in the food and drug industry.
The authors of this article consider it necessary to thank and appreciate the research deputy of Khuzestan Agricultural Research Center and the management of Parham South Company who collaborated with us in this study.
Authors declared no conflict of interests.
Received: 2019/08/8 | Accepted: 2019/11/30 | ePublished: 2020/01/10
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |






