BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-750-en.html
2- Department of Microbiology, Saveh Branch, Islamic Azad University, Saveh, Iran
.
Shigella species are members of the Enterobacteriaceae family with four groups: group A (Shigella dysentery), group B (Shigella flexneri), group C (Shigella boydii) and group D (Shigella sonnei) (1). The infectious dose of Shigella is less than 200 bacilli; therefore, shigellosis can spread rapidly among communities with low health standards (2). Bacterial dysentery (Bacillary dysentery or shigellosis) is considered as one of the most important acute gastrointestinal diseases, especially in children and immune-compromised patients which cause high number of fatalities in addition to economic losses and social problems (3). Anorexia, fever, intestinal inflammation, bloody-purulent stools, abdominal pain, tenesmus, and feeling of incomplete bowel emptying with anal pain are the symptoms of this disease (4). It is estimated that 165 million cases of bacillary dysentery and 1.1 million deaths occur annually worldwide, about 60% of which are in children younger than five years old (5).
The appropriate and timely antibiotic treatment shortens the course of the disease and reduces its complications and prevents the transmission of the disease to the healthy people. Today, due to the widespread use of antibiotics, Shigella species have become resistant to many antibiotics, including the third generation of cephalosporins. This issue has made the treatment of this disease difficult (6). The most important problem in the treatment of people with shigellosis is the development of multi-drug resistance (MDR) (7). Production of the beta-lactamase enzymes is the main reason for the gram-negative bacteria resistance to beta-lactams (8). These enzymes destroy the amide ring of beta-lactams. Beta-lactamase enzymes are divided into four groups A to D based on the amino acid sequences. Broad-spectrum beta-lactamase enzymes (ESBLs) are from class A beta-lactamases that hydrolyze broad-spectrum cephalosporins with a side chain of Oximino, causing bacterial resistance to penicillins, the first, second and third generations of cephalosporins and aztreonam. They are inhibited by beta-lactamase inhibitors such as clavulanic acid (9).
The CTX-M beta-lactamase genes were first reported in 1989 from Japan and spread to the other parts of the world since then (10). Today, these beta-lactamase enzymes are divided into five groups: CTX-M1 was reported from Germany in 1989 (11), CTX-M-2 was reported from Japan in 1986, CTX-M-8 was reported from Brazil in 1996 (12), CTX-M-9 was reported from Spain in 1994, and the CTX-M-25 was reported from Canada in 2000 (10). These beta-lactamase genes have little association with the TEM and SHV beta-lactamase members, and instead there is a high similarity between the chromosomal enzymes Amp-C, especially KLU-1 and KLU2, and the CTX-M enzymes. There are theories based on the derivation of these enzymes from one species (13).
Broad-spectrum beta-lactamase-producing bacteria are placed in the human and animal intestines which are very difficult to control and eradicate because ESBL genes can be transmitted between different genus, species, and different strains of intestinal bacteria (7). There are some reports of Shigella sonnei multiple-resistance to antibiotics, especially cephalosporins third-generation such as cefotaxime and ceftriaxone.
In recent years, the emergence of the antibiotic resistance phenomenon has raised many concerns in the medical communities due to the failure of the treatment process (8). Due to the development of broad-spectrum beta-lactam antibiotic resistance genes in Shigella sonnei strains and the high speed and accurate detection of molecular methods and simultaneous identification of several genes, this study aimed to identify broad-spectrum beta-lactamase genes CTX-M-2, CTX-M and Ampc-dependent CMY gene in Shigella sonnei strains isolated from diarrheal specimens by Multiplex-PCR method and the antibiotic resistance pattern of these strains.
Finally, to differentiate and confirm Shigella species the biochemical tests (oxidase, catalase, SIM, MRVP, citrate consumption, TSI test, urease production, phenylalaninidaminase, lysine decarboxylase, sodium malonate, and decarboxylation of the amino acids ornithine and mannitol) were conducted. The isolates with biochemical properties: lactose negative, gas production negative, immobilized, lysine decarboxylation negative, citrate negative, urea hydrolysis negative and methyl red positive were considered as Shigella isolate. Serotyping tests were performed on the slides of fresh Shigella cultures using Baharafshan Co. kits by agglutination method (1, 2).
Disk Diffusion
The antibiotic susceptibility test was performed using disk diffusion method and according to the instructions of the Laboratory and Clinical Standards Institute (14) on Müller-Hinton agar medium (Merck, Germany) for the antibiotics trimethoprim-sulfamethoxazole (25 µg), erythromycin (30 µg), amoxicillin (25 µg), ceftriaxone (30 µg), cefepime (30 µg), amoxiclav (25 µg) (produced by Himedia Co, India).
After bacterial DNA extraction using gram-negative bacterial genomic DNA extraction kit of Iranian Biological Resource Center (Cat no. MBK0041) and confirmation of the extracted DNA purity using biophotometer (Bio-Rad, USA), the genes CTX-M-2, CTX-M-8 and CMY were amplified using M-PCR method and specific primer sequences (Table 1) in thermocycler (Eppendorf, Germany) in the final volume of 25 µL including 10.5 µL PCR master mix 5X (Sinaclon, Iran) containing Taq DNA polymerase (0.05 U/µL), MgCl2 (3 mM) and dNTPs (0.4 mM), 0.7 μL of each primer, 1 μL of template DNA (10 ng) and 12.1 μL of double-distilled water for 33 cycles. The primer BLAST was performed on the website https://www.ncbi.nlm.nih.gov/tools/primer-blast/.
Thermal cycling included denaturation step at 95°C for 1 min, primer annealing at 60°C for 30 sec and elongation step at 72°C for 1 min. Finally, the PCR products were run on 1% agarose gel containing ethidium bromide ( Figure1). In the molecular study, Shigella sonnei ATCC 25931 and Escherichia coli ATCC 25922 (prepared from the Microbial Bank of Pasteur Institute of Iran) were used as negative and positive controls, respectively.
Statistical Analysis
Data were collected and analyzed using SPSS software version 16. The statistical analysis was performed using Pearson-Chi Square test. The P-value<0.05 was considered as significant level.
Table 1. Sequence of the primers
| Target gene | Primer sequence (3´ to 5´) | Product length (bp) |
| CTX-M-2 | F: TTAATGATGACTCAGAGCATTC R: GATACCTCGCTCCATTTATTG |
901 |
| CTX-M-8 | F: CGCTTTGCCATGTGCAGCACC R: GCTCAGTACGATCGAGCC |
307 |
| CMY | F: TGGCCAGAACTGACAGGCAAA R: TTTCTCCTGAACGTGGCTGGC |
462 |
| Antibiotic | Sensitive (S) (%) | Intermediate (I) (%) | Resistant (R) (%) |
| Cotrimoxazole | 60 (100) | 0 (0.0) | 0 (0.0) |
| Erythromycin | 30 (50) | 4 (6.7) | 26 (43.3) |
| Amoxicillin clavulanic acid | 40 (66.6) | 8 (13.4) | 12 (20) |
| Cefepime | 30 (50) | 6 (10) | 24 (40) |
| ceftriaxone | 40 (86.6) | 0 (0.0) | 8 (13.4) |
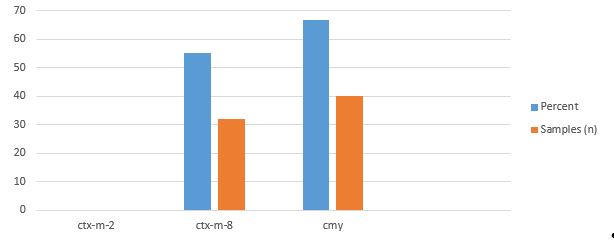
The presence or absence of broad-spectrum beta-lactamase genes was studied on all 60 Shigella sonnei isolates of the children's diarrheal stool samples. The frequencies of CMY and CTX-M-8 genes were 66.7% (n=40) and 55% (n=33), respectively ( Figure1). All the strains were negative regarding CTX-M-2 gene ( Figure 2).
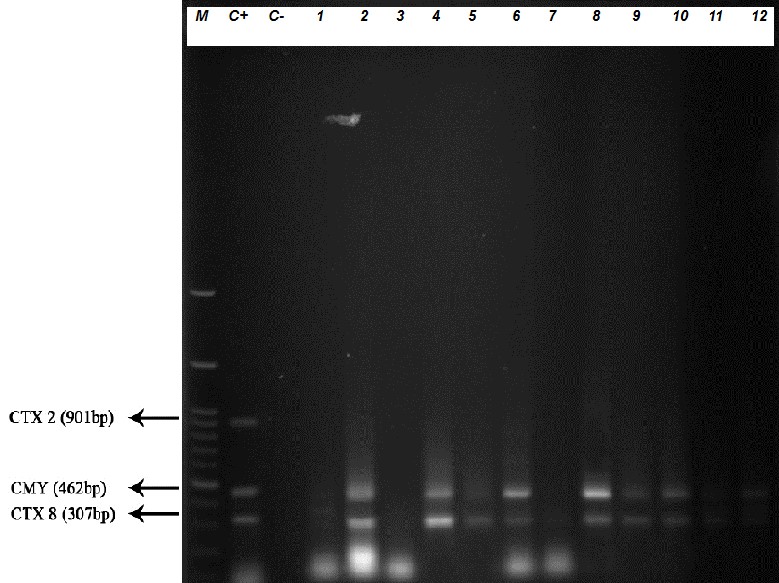
Figure 1. The M-PCR test result on some isolates. C+: Positive control (E. coli ATCC 25922), C: Negative control (Shigella sonnei ATCC 25931). The numbers 1-12 are clinical strains collected from diarrhea (M: DNA marker: 100 bp, Fermentas).
Figure 2. Number and results of beta-lactamase genes in Sony Shigella samples
Shigellosis is endemic worldwide and is the most common cause of bacterial dysentery (1). The disease heals spontaneously in adults, while it is very dangerous in infants and children which can lead to death (3). In developing countries, this disease still remains an important health issue (4).
Due to the increase in antibiotic resistance in intestinal pathogens, it seems that opting a suitable drug to treat these infections has become difficult, thus, determining the drug resistance pattern on a regional and periodic basis seems necessary for this bacterium. In accordance with the present study, Abbaspour et al., (16) in 2014 showed that 23.3% and 24.4% of Shigella sonnei isolates were resistant to ceftriaxone and cefixime, respectively. In the present study, 31.6% of the strains were MDR and the highest resistance (40%) was related to cefepime. The results showed a significant association (P<0.02) between the presence of ESBL genes and the incidence of resistance to cephalosporins such as cefepime. In a study conducted by Bonnet et al., in 2007 in Brazil, the beta-lactamase genes CMY-2 and AmpC were examined. In line with our study, all the strains with multiple resistance in the Bonnet et al., study contained CMY gene (17).
Like our study, Hussain et al., in Pakistan in 2011 showed that the frequencies of CTX-M and CMY genes were 57.7% and 50%, respectively (18). Contrary to the present study, Mendonça et al., in 2007 in Portugal showed that blaCTX-M-2 had the highest abundance among other genes in the isolates (19). This difference can be due to differences in the year of the study, geographical distance, variety of strains and sample size. Again, contrary to the present study, the broad-spectrum beta-lactamase gene CTX-M-2 in cefotaxime-resistant Shigella sonnei was detected in Argentina by molecular method in the study conducted by Radice et al., in 2001. The strains containing this gene were reported to be resistant to commonly used cephalosporins (20). This gene was not detected in any of the samples in our study. This difference could be due to the geographical distance and genetics of the strains.
More comprehensive studies are needed to identify different types of beta-lactamase classes and other resistance genes in Shigella strains in the country. Also, by determining the antibiotic resistance pattern and using the appropriate antibiotics when treatment is needed, antibiotic resistance and the spread of resistant strains in the community among human populations can be prevented. One of the salient features of the present study is the simultaneous study of resistance genes using M-PCR method. It is suggested that, the presence of other resistance genes and determining the genetic relationship of these strains should be considered in the future studies.
Based on the present study results, it was found that the highest resistance in Shigella sonnei strains was against cefepime and the most common broad-spectrum beta-lactamase gene in these strains was CMY. There was a statistically significant association between the presence of CMY gene and resistance to cefepime (P<0.05). For the high prevalence of beta-lactam antibiotic resistance genes in Shigella sonnei strains, careful medical care and proper and timely use of appropriate antibiotics are essential to prevent the spread of resistant strains.
The authors would like to thank Dr. Fereshteh Shahcheraghi for providing positive control strain. We also thank the staff of the Children's Medical Center (Tehran) for the preparation of microbial strains and the management and the staff of the Pasargad Research Laboratory who assisted in carrying out this project.
Authors declared no conflict of interests.
Received: 2017/08/26 | Accepted: 2017/10/21 | ePublished: 2020/10/5
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |