BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-2520-en.html
2- Departement of Biochemistry, Faculty of Medicine, University of Palangka Raya, Palangkaraya, Indonesia
3- Departement of Parasitology, Faculty of Medicine, University of Palangka Raya, Palangkaraya, Indonesia
Infectious diseases are a major health concern, and high humidity levels can hasten their development particularly in tropical areas (1). Infectious diseases are caused by microorganisms such as bacteria, viruses, fungi, or parasites. Certain bacteria, such as S. aureus, P. aeruginosa, and S. pyogenes, play a role in causing infectious diseases (2-4).
Staphylococcus aureus is a prevalent opportunistic pathogen in humans, responsible for various illnesses. These range from minor skin infections like pimples and boils to severe conditions such as sepsis, pneumonia, endocarditis, and osteomyelitis (5). Pseudomonas aeruginosa, a Gram-negative opportunistic pathogen, infects patients with conditions such as cystic fibrosis, chronic obstructive pulmonary disease (COPD), cancer, and severe infections requiring ventilation, including those caused by COVID-19. It is also known for its multi-drug resistance (3, 6, 7). Streptococcus pyogenes, also known as group A streptococcus (GAS), causes various human diseases. These include strep throat (pharyngitis), skin or wound infections (such as impetigo), and scarlet fever (8).
The prevalence of bacterial diseases is rising because these microorganisms are becoming resistant to different antibiotics. For instance, methicillin-resistant S. aureus (MRSA) is now a significant source of infections in both healthcare facilities and community settings (5). Antimicrobial resistance poses a significant risk to global health systems by undermining the effectiveness of preventing and treating infections caused by bacteria. Currently, around 700,000 people worldwide die annually due to drug-resistant infections. Projections suggest that by 2050, the mortality rate attributed to antimicrobial resistance will rise to 4.73 million in Asia, 4.15 million in Africa, 0.39 million in Europe, 0.392 million in Latin America, 0.317 million in North America, and 0.022 million in Oceania (9).
Global health faces a significant challenge due to the rise of antibiotic resistance. Consequently, there's a demand for innovative treatments to replace ineffective antimicrobial medications. Streptomyces, a prolific producer of natural products, contributes significantly to the pool of antimicrobial agents (10, 11). In a study conducted in Thailand, researchers isolated and identified bioactive compounds from Streptomyces actinomycinicus (S. actinomycinicus) PJ85. Among these compounds, PJ85_F39 was identified as dihomo-linolenic acid (DGLA). Notably, this study represents the first report on the purification and characterization of antibacterial compounds from S. actinomycinicus. Given the limited natural source of DGLA, S. actinomycinicus PJ85 could serve as a valuable reservoir for DGLA production, potentially playing a significant role in the pharmaceutical industry (12).
Dihomo-γ-linolenic acid (DGLA) is an omega-6 polyunsaturated fatty acid with 20 carbon atoms and three double bonds. It serves as a precursor for the biosynthesis of arachidonic acid (ARA) and has been associated with antimicrobial, anti-inflammatory, and antiallergic properties (13, 14). In 2013, Desbois and Lawlor (15) documented that dihomo-γ-linolenic acid (DGLA) exhibited antibacterial effects against gram-positive bacteria, including Propionibacterium acnes and S. aureus, with minimum inhibitory concentration (MIC) values of 128 mg/L and 1024 mg/L, respectively (15). Earlier studies have acknowledged that soil fungi from the Mortierella genus-such as M. alpina, M. clonocystis, M. elongate, M. gamsii, M. humilis, M. macrocystis, and M. globulifera are proficient at producing dihomo-γ-linolenic acid (DGLA) (16).
Despite the strides made in antimicrobial therapies, the rapid emergence of antibiotic-resistant bacteria has severely compromised the effectiveness of existing treatments. Methicillin-resistant S. aureus (MRSA), multi-drug-resistant P. aeruginosa, and drug-resistant S. pyogenes exemplify the growing challenge of treating bacterial infections. The development of resistance mechanisms by these pathogens, such as the production of beta-lactamase enzymes, efflux pumps, and biofilm formation, undermines the efficacy of conventional antibiotics and necessitates the exploration of alternative and innovative therapeutic strategies.
Current antimicrobial therapies often fail to target the specific virulence factors of bacteria, which play a crucial role in their pathogenicity and resistance. Additionally, the potential for adverse effects, reduced potency over time, and the narrow spectrum of activity of some antibiotics further limit their utility. Therefore, there is an urgent need for novel antimicrobial agents that can effectively inhibit or neutralize bacterial virulence factors without contributing to resistance.
This study aims to fill this gap by investigating the interaction of dihomo-γ-linolenic acid (DGLA) with the virulence factors of S. aureus, P. aeruginosa, and S. pyogenes. By employing a comprehensive bioinformatics approach, the research seeks to elucidate the specific protein-compound interactions that confer antibacterial properties to DGLA. The findings of this study represent the first evidence of DGLA's efficacy against drug-resistant microorganisms, providing a promising avenue for developing new antimicrobial therapies that mitigate the limitations of current treatments.
While DGLA has been documented to exhibit antibacterial properties, there is currently no evidence supporting its effectiveness against MRSA. Consequently, this study represents the first evidence that DGLA is also efficacious against drug-resistant microorganisms.
The selection of S. aureus, P. aeruginosa, and S. pyogenes for this study is based on their significant clinical relevance and their roles as major contributors to infectious diseases. Staphylococcus aureus is a prevalent opportunistic pathogen responsible for a wide range of illnesses, from minor skin infections to severe conditions such as sepsis and pneumonia. Its capacity to develop resistance to multiple antibiotics, including methicillin-resistant S. aureus (MRSA), poses a substantial threat to public health.
Pseudomonas aeruginosa, a Gram-negative opportunistic pathogen, is notorious for infecting patients with chronic conditions such as cystic fibrosis and chronic obstructive pulmonary disease (COPD), as well as those with weakened immune systems. Its ability to resist multiple drugs complicates treatment, making it a critical target for novel antimicrobial agents.
Streptococcus pyogenes, also known as group A streptococcus (GAS), causes various human diseases, including strep throat, impetigo, and scarlet fever. Its role in both mild and life-threatening infections, coupled with the emergence of drug-resistant strains, underscores the importance of exploring new therapeutic options.
By focusing on these three bacterial strains, the study aims to address the pressing need for effective treatments against some of the most challenging and resistant pathogens in clinical settings.
2.1 Protein Interaction Analysis
This study conducted bioinformatics research using FASTA samples from S. aureus, P. aeruginosa, and S. pyogenes obtained from the National Center for Biotechnology Information (NCBI). The research focused on examining the virulence factors of S. aureus, P. aeruginosa, and S. pyogenes that are affected by DGLA.
STITCH v5.0 was used to analyze protein-compound interactions. The STITCH database is available for full download, programmatically accessible through an extensive API, or searchable via a redesigned web interface at http://stitch.embl.de (17).
2.2. Molecular Docking
Molecular Docking Analysis
To structurally validate the predicted chemical-protein interactions from the STITCH database, molecular docking studies were performed for selected bacterial proteins with Dihomo-γ-linolenic Acid (DGLA). Proteins were selected based on their interaction confidence scores, known functional relevance, and biological significance. The Epidermin leader peptide processing serine protease (EpiP) from S. pyogenes, Acyl-CoA Thioesterase (tesB; PDB ID: 5V10) from P. aeruginosa PAO1, and ScpA15 from S. pyogenes were selected for detailed molecular docking analysis based on their relevance in bacterial virulence and lipid metabolism.
Protein structures were retrieved from the Protein Data Bank (PDB). The structures were processed by removing solvent molecules, adding polar hydrogens, and assigning Gasteiger charges using AutoDockTools (ADT). The ligand structure (DGLA) was obtained from the PubChem database and optimized similarly using ADT for proper torsional flexibility. Docking was performed using AutoDock Vina, employing a grid box sized and centered to encompass the ligand-binding site of each protein fully. Blind docking approaches were also performed for proteins where binding site prediction was challenging. Docking calculations were conducted with an exhaustiveness level of 8 to ensure accurate and reliable sampling of potential ligand conformations.
The binding affinity, RMSD, and number of hydrogen bonds were evaluated to select the most favorable ligand-protein complexes. Docking poses were further analyzed using UCSF ChimeraX to assess the presence of hydrogen bonds, π-stacking interactions, and salt bridges, providing deeper insight into ligand stability and specificity within the protein active sites. Validation of docking results considered the binding energies, number of hydrogen bonds, and biological plausibility of interactions. The final candidate poses were those demonstrating the most robust binding energies and stable ligand-protein interactions, confirming the predicted functional associations suggested by STITCH database analysis.
References for the software used:
-
AutoDock Vina (Trott & Olson, 2010) docking
-
ChimeraX (UCSF ChimeraX)
-
OpenBabel (file format conversion)
2.3 Functional Classification
The proteins targeted by DGLA from S. aureus, P. aeruginosa, and S. pyogenes were examined for their functional class using VICMPred. The web server VICMpred has been developed and can be accessed at http://www.imtech.res.in/raghava/vicmpred/ (18).
2.4 Virulence Prediction
The virulence properties of these proteins were assessed using VirulentPred. VirulentPred 2.0 is available as a user‐friendly web interface at https://bioinfo.icgeb.res.in/virulent2/ and a standalone application suitable for bulk predictions (19).
VICMpred and VirulentPred, both utilizing an SVM-based approach, employ dipeptide composition, amino acid composition, and other patterns to predict virulence factors. Their accuracy ranges from 70% to 80% (18, 20). VICMpred is a web server that broadly classifies bacterial proteins into functional categories, including information molecules, cellular processes, virulence factors, and metabolism molecules. In contrast, VirulentPred focuses solely on distinguishing proteins as either virulent or non-virulent.
2.5 Additional Analyses
Epitope B cell analysis employed BepiPred v.2. Furthermore, PSORTb v.3 was utilized for subcellular protein localization analysis.
Several HPs that show virulent results with VirulentPred will be checked for classification of protein families with InterproScan. InterPro provides functional analysis of proteins by classifying them into families and predicting domains and important sites. To classify proteins in this way, InterPro uses predictive models, known as signatures, provided by several different databases. The InterPro database (https://www.ebi.ac.uk/interpro/) provides an integrative classification of protein sequences into families and identifies functionally important domains and conserved sites (21).
2.6 Parameters Used in Each Tool and Justify
STITCH v5.0 was configured with default parameters, which include a confidence score cutoff of 0.7 to ensure high reliability of predicted interactions. This score threshold was chosen based on its balance between stringency and inclusivity, allowing for the identification of significant protein-compound interactions while minimizing false positives (17).
For VICMPred, the tool's default settings were employed, which leverage dipeptide and amino acid composition to classify bacterial proteins. The accuracy of VICMPred, ranging from 70% to 80%, is a testament to its robust predictive capability, thereby justifying its use for functional classification in this study. The choice of VICMPred is supported by prior studies (20) which highlight its efficiency and reliability in categorizing bacterial proteins.
VirulentPred was utilized with its default parameter settings, which involve an SVM-based approach incorporating dipeptide composition and amino acid composition patterns. This methodology has been validated in various studies (20) for its effectiveness in predicting virulence factors with commendable accuracy. The decision to use VirulentPred stems from its proven track record in virulence factor prediction and its user-friendly interface that facilitates bulk predictions.
Epitope B cell analysis was conducted using BepiPred v.2, with thresholds set according to the tool’s recommendations, ensuring optimal sensitivity and specificity (22). PSORTb v.3 was utilized for subcellular localization with its default settings, which have been validated for high precision in bacterial protein localization studies (23).
For classification of protein families, InterProScan was employed, utilizing predictive models from various databases integrated into InterPro. The signatures for these models have been curated to provide comprehensive functional analysis (24). InterProScan was selected due to its extensive database support and high accuracy in domain and site prediction.
3.1 Protein Interaction Analysis
The analysis of protein interactions was performed using STITCH v5.0. The results revealed that several proteins from P. aeruginosa, S. aureus, and S. pyogenes interacted with DGLA. Furthermore, these proteins were also found to interact with each other (Figure 1).
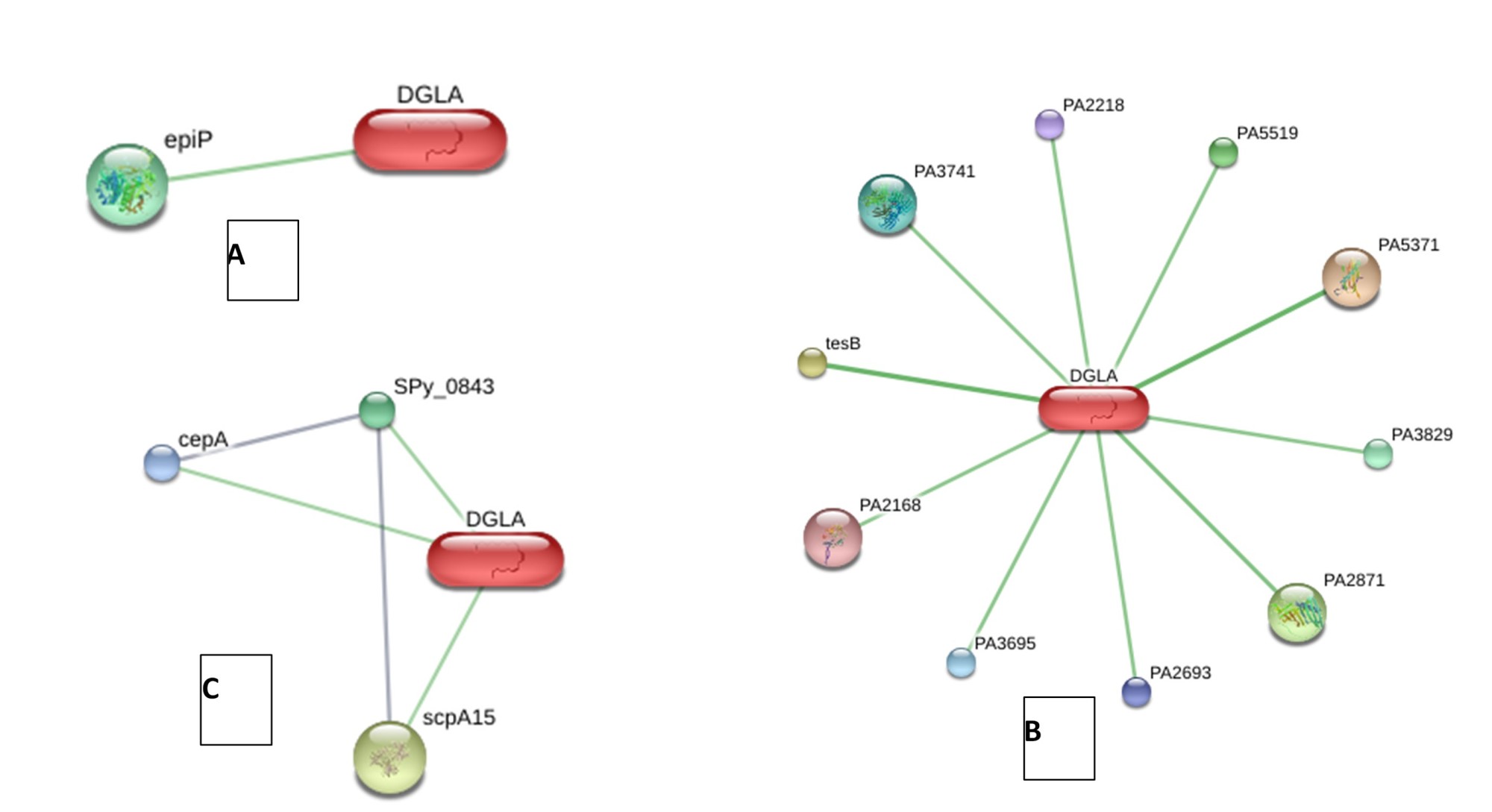
Figure 1. Protein Interaction Analysis using STITCH v5.0
A: Interaction Analysis of S. aureus COL with DGLA; Predicted Functional Partners: epidermin leader peptide processing serine protease (epiP)
B: Interaction Analysis of P. aeruginosa with DGLA; 9 hypothetical proteins: PA5371, PA2871, PA5519, PA3829, PA3741, PA3695, PA2693, PA2218, PA2168. acyl-CoA thioesterase (tesB)
C: Interaction Analysis of S. pyogenes with DGLA; C5A peptidase (scpA15), cell envelope proteinase (cepA), SPy_0843 (hypothetical protein)
3.2 Molecular Docking Simulations
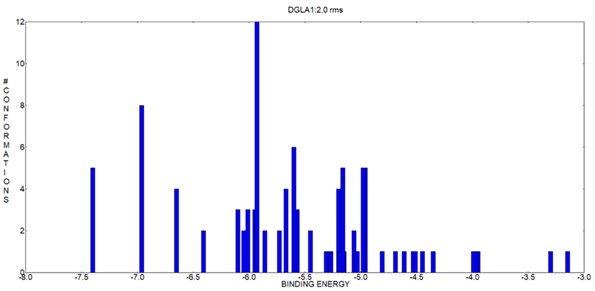
The molecular docking results demonstrated that DGLA exhibited a strong binding affinity towards the selected bacterial proteins. The binding energy values for the best-ranked docking poses were as follows: TesB (-7.4 kcal/mol), ScpA (-4.3 kcal/mol), and EpiP (-4.1 kcal/mol). The docking scores suggest that DGLA had the highest affinity towards TesB from P. aeruginosa, indicating potential inhibition of its enzymatic function (Table 1).
TesB-DGLA interaction: The lowest energy pose (-7.4 kcal/mol) showed strong van der Waals and hydrogen bonding interactions with VAL-33, ASP-21, and TYR-32 (Figures 2-5). The hydrogen bond distance between DGLA and active site residues ranged from 2.1 to 3.5 Å, confirming stable binding.
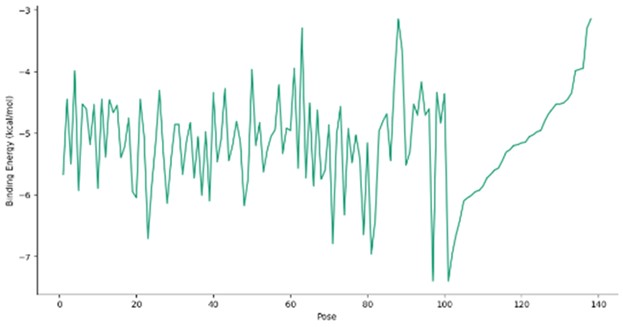
Figure 2. Binding energy profile for the docking poses of P. aeruginosa Acyl-CoA thioesterase (PDB ID: 5V10) with DGLA. The x-axis represents different docking poses, while the y-axis denotes the binding energy (kcal/mol).
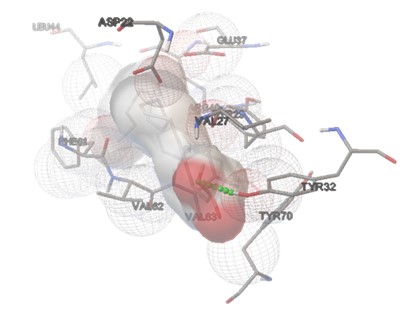
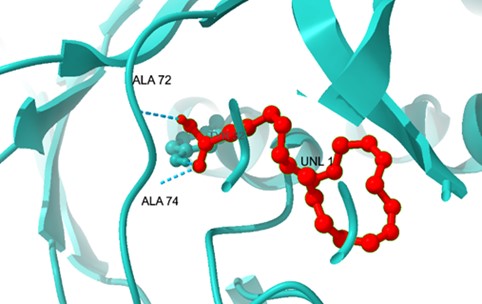
Figure 3. Left: The top docking pose of DGLA within the active site of P. aeruginosa Acyl-CoA thioesterase (PDB ID: 5V10). The ligand is shown in red sticks, and interacting residues are labeled. Hydrogen bonds are indicated with blue dashed lines. Right: Molecular interaction surface representation of DGLA binding to the active site of P. aeruginosa Acyl-CoA thioesterase. The protein is shown in a molecular surface representation, and the ligand is embedded in the pocket.
Figure 4. Hydrogen bonding and hydrophobic interactions between DGLA and key residues in P. aeruginosa Acyl-CoA thioesterase (5V10). The molecular surface around the ligand is displayed, with interacting residues highlighted.
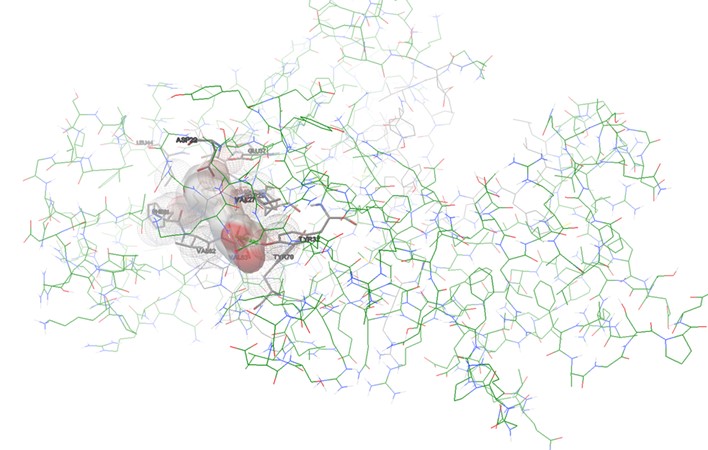
Figure 5. The 2D ligand interaction diagram for DGLA docked with P. aeruginosa Acyl-CoA thioesterase (5V10). Hydrogen bond interactions are represented in green, and hydrophobic interactions are in purple.
Table 1. Docking results of P. aeruginosa Acyl-CoA thioesterase (PDB ID: 5V10) with DGLA.
| Cluster Rank | Binding Energy (kcal/mol) | Estimated Inhibition Constant Ki (µM) | Intermolecular Energy (kcal/mol) | vdW + Hbond + Desolvation Energy (kcal/mol) | Electrostatic Energy (kcal/mol) | Total Internal Energy (kcal/mol) | Torsional Energy (kcal/mol) | Unbound System Energy (kcal/mol) |
| 97 | -7.40 | 3.77 | -12.17 | -12.04 | -0.14 | -0.94 | 4.77 | -0.94 |
| 71 | -6.96 | 7.89 | -11.73 | -11.31 | -0.42 | -1.79 | 4.77 | -1.79 |
| 74 | -6.65 | 13.36 | -11.42 | -11.37 | -0.06 | -1.34 | 4.77 | -1.34 |
| 28 | -6.41 | 19.96 | -11.18 | -11.07 | -0.12 | -1.05 | 4.77 | -1.05 |
| 29 | -6.10 | 33.57 | -10.88 | -10.80 | -0.08 | -1.21 | 4.77 | -1.21 |
ScpA-DGLA interaction: The top docking pose (-4.3 kcal/mol) exhibited moderate affinity and positioned DGLA within the substrate-binding pocket of ScpA. Key interactions were observed with Lys-185 and Asp-140, forming hydrogen bonds at 2.5 Å and 2.9 Å, respectively (Figure 6).
EpiP-DGLA interaction: The docking of DGLA to EpiP resulted in a binding energy of -4.1 kcal/mol, indicating weaker interactions compared to TesB. However, hydrophobic contacts with Ala-72 and Ala-74 contributed to ligand stabilization within the active pocket (Figure 7).
The binding energy distribution across docking poses is illustrated in Figure 1, revealing variability in ligand orientations and interaction strengths. The 3D visualization of docking complexes (Figures 5-7) provides insights into specific residue interactions and ligand conformations within the binding pocket.
These findings suggest that TesB represents the most promising target for DGLA binding, while ScpA and EpiP interactions indicate potential but weaker binding affinities. Further experimental validation, including enzyme inhibition assays and molecular dynamics simulations, will be necessary to confirm these in silico predictions.
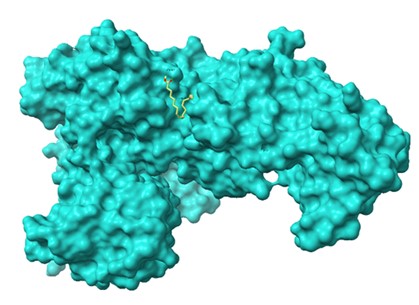
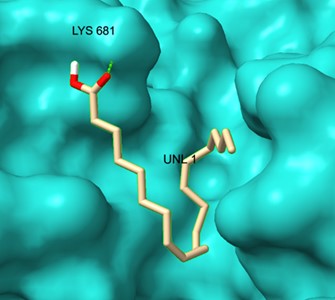
Figure 6. Molecular docking analysis of DGLA with ScpA (PDB: 7YZX) showing key active site interactions.

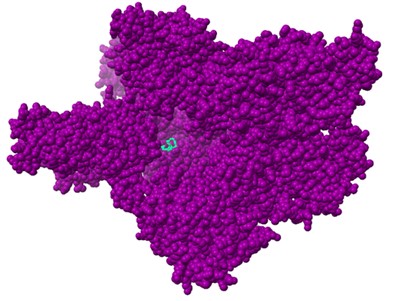
Figure 7. Binding conformation of DGLA in the EpiP protein with major interacting residues.
3.3 Functional Class and Virulence Property Analysis of Proteins
Following the protein interaction analysis, VICMPred and VirulentPred were employed to assess functional classes and virulence properties for each protein (Tables 2 and 3).
Table 2. Functional Class and Virulence Property Analysis of organism Proteins Interacting with DGLA
| Organism | Identifier | Protein Interacts DGLA |
VICMPred Functional Class | VirulentPred |
| S. aureus COL | epiP | epidermin leader peptide processing serine protease | Virulence factors | Virulent |
P. aeruginosa |
PA5371 | hypothetical protein | Cellular process | Virulent |
| tesB | acyl-CoA thioesterase | Metabolism Molecule | Virulent | |
| PA2871 | hypothetical protein | Metabolism Molecule | Non- Virulent | |
| PA5519 | hypothetical protein | Cellular process | Non- Virulent | |
| PA3829 | hypothetical protein | Metabolism Molecule | Virulent | |
| PA3741 | hypothetical protein | Cellular process | Virulent | |
| PA3695 | hypothetical protein | Metabolism Molecule | Non- Virulent | |
| PA2693 | hypothetical protein | Cellular process | Non- Virulent | |
| PA2218 | hypothetical protein | Metabolism Molecule | Non- Virulent | |
| PA2168 | hypothetical protein | Cellular process | Virulent | |
S. pyogenes |
scpA15 | C5A peptidase | Metabolism Molecule | Virulent |
| SPy_0843 | hypothetical protein | Virulence factors | Virulent | |
| cepA | cell envelope proteinase | Virulence factors | Virulent |
Table 3. Virulent Hypothetical Proteins classification with InterPro
| Organism | Hypothetical Proteins (HPs) Identifier | Protein family membership InterPro |
P. aeruginosa |
PA5371 | Cytosolic acyl coenzyme A thioester hydrolase |
| PA3829 | FrsA esterase | |
| PA3741 | Cytosolic acyl coenzyme A thioester hydrolase | |
| PA2168 | FrsA esterase | |
| S. pyogenes | SPy_0843 | Putative surface bspA-like protein |
3.4 B-cell Epitope Analysis
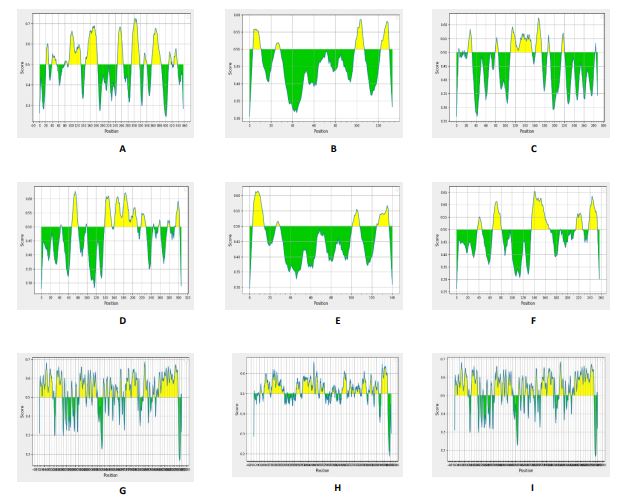
This analysis used BepiPred v2.0, focusing on virulent proteins obtained from examination using VirulentPred (Figure 8).
3.5 Subcellular Location Analysis
The analysis employed PSORTb version 3.0 to identify proteins with virulent traits.” PSORTb v3.0 is a precise bacterial localization prediction tool that handles both archaeal and bacterial sequences (25).
Figure 8. Results of B-cell Epitope Analysis on virulent proteins of S. aureus, P. aeruginosa, and S. pyogenes.
A : Analysis of S. aureus COL; leader peptide processing serine protease (epiP)
B-F : Analysis of P. aeruginosa; (B) PA5371, (C) tesB, (D) PA3829, (E) PA3741, (F) PA2168.
G-I : Analysis of S. pyogenes; (G) scpA15, (H) SPy_0843, (I) cepA
Table 4. Subcellular Location Analysis of Virulent organism Proteins Interacting with DGLA
| Organism | Identifier | Functional Proteins | Subcellular Location |
| S. aureus COL | epiP | epidermin leader peptide processing serine protease | Extracellular |
P. aeruginosa |
PA5371 | Cytosolic acyl coenzyme A thioester hydrolase | Unknown |
| tesB | acyl-CoA thioesterase | Cytoplasmic | |
| PA3829 | FrsA esterase | Cytoplasmic | |
| PA3741 | Cytosolic acyl coenzyme A thioester hydrolase | Unknown | |
| PA2168 | FrsA esterase | Cytoplasmic | |
| S. pyogenes | scpA15 | C5A peptidase | Cellwall |
| SPy_0843 | Putative surface bspA-like protein | Cellwall | |
| cepA | cell envelope proteinase | Cellwall |
In recent years, bioinformatics has become a pivotal tool in the discovery of antimicrobial agents, leveraging computational techniques to predict and analyze interactions between drugs and microbial targets. Previous studies, such as those by Altschul et al (26) and Kanehisa et al (27) have demonstrated the potential of bioinformatics in identifying novel drug targets by comparing protein sequences and functions across different pathogens. These studies have laid the groundwork for understanding the complex interactions within microbial communities and have provided valuable insights into the mechanisms of action of various antimicrobial agents.
Based on the findings, STITCH v5.0 revealed that a protein from S. aureus, specifically epiP, interacts with DGLA. Additionally, P. aeruginosa has ten proteins (PA5371, tesB, PA2871, PA5519, PA3829, PA3741, PA3695, PA2693, PA2218, and PA2168), while S. pyogenes has three proteins (scpA15, SPy_0843, and cepA) that also interact (Figure 1). These proteins were mentioned in various forms of identification codes (identifier). In the interaction between DGLA and proteins from S. pyogenes, there appears to be cross interaction between the proteins (Figure 1-C). Protein-protein interactions play a crucial role in aiding cells to perform their biological functions (28, 29).
In the field of bioinformatics, protein interactions targeted by DGLA were identified based on similarities in their amino acid sequences, some of which also shared similar functions. These interactions, discovered through bioinformatics methods, can enhance our understanding beyond in vitro studies. For instance, a study by Chanthasena et al (12) found that S. actinomycinicus PJ85 might serve as a potent source of DGLA due to its limited natural availability and antibacterial activity. While the precise bacterial component responsible for interacting with DGLA and causing growth inhibition remains unknown, this study has successfully identified the specific proteins that directly interact with DGLA.
The majority of proteins that interact with DGLA in S. aureus, P. aeruginosa, and S. pyogenes play essential roles in their survival. Functional class and virulence property analysis using VICMPred revealed that these proteins are involved in term "cellular process" and "metabolism molecule". Cellular processes such as cell division, membrane biogenesis, cell movement, and signal transduction. Additionally, metabolism molecule, including energy production, transportation, and the metabolism of carbohydrates, amino acids, nucleotides, and lipids (30).
In various organisms, over 30% of proteins have unknown molecular functions and are referred to as 'hypothetical proteins' (HPs). Annotating these proteins functionally can provide insights into their roles in different metabolic processes and help identify novel drug targets (31).
Various bioinformatics resources, including databases and tools, exist for annotating the functions of hypothetical proteins. These resources have proven effective in characterizing the roles of such proteins in diverse bacterial pathogens (32, 33).
A hypothetical protein is a predicted protein encoded by a known open reading frame, but its specific function remains unknown due to a lack of experimental evidence. In most genomes, approximately half of the protein-encoding genes fall into the category of hypothetical proteins (HPs). These HPs likely play a significant role in an organism’s overall proteomic landscape. Accurate annotation of HPs in pathogens enhances our understanding of virulence mechanisms, facilitates the discovery of new protein structures and pathways, and identifies potential drug targets. To classify an HP as an essential gene and a novel drug target, it must be pathogen-specific (non-homologous to the host) and involved in critical pathogen processes such as replication, survival, virulence, or growth (30). In this study, several 'hypothetical proteins' (HPs) were obtained, that is 9 HPs (PA5371, PA2871, PA5519, PA3829, PA3741, PA3695, PA2693, PA2218, and PA2168) from P. aeruginosa and 1 HPs (SPy_0843) from S. pyogenes.
The interaction of DGLA with specific proteins in S. aureus, P. aeruginosa, and S. pyogenes opens up promising avenues for therapeutic interventions. DGLA, a polyunsaturated fatty acid, has been observed in previous studies to possess anti-inflammatory and antimicrobial properties (34, 35). The findings in this study highlight its role in modulating proteins that are critical to the survival and virulence of these pathogens.
Several mechanisms could be proposed for how DGLA modulates virulence factors. For instance, DGLA has been shown to integrate into bacterial membranes, altering their fluidity and permeability, which can disrupt membrane-bound enzymes and transport systems essential for bacterial virulence. Additionally, DGLA's ability to produce reactive oxygen species (ROS) can lead to oxidative stress within bacterial cells, impairing their metabolic functions and reducing their pathogenicity (36).
In the case of S. aureus, the interaction between DGLA and the epiP protein suggests potential therapeutic applications. EpiP is involved in processing lantibiotics, which have strong antibacterial properties. By modulating epiP, DGLA could enhance the production of these lantibiotics, thereby bolstering the body's natural defenses against MRSA infections (5, 37).
For P. aeruginosa, the identified HPs, such as PA5371 and tesB, play roles in biofilm formation and antibiotic resistance. The modulation of these proteins by DGLA could inhibit biofilm formation, making the bacteria more susceptible to antibiotic treatments (38).
Furthermore, the esterase activity of FrsA family proteins, disrupted by DGLA, could impair the pathogen's ability to breakdown essential compounds, reducing its virulence and growth.
Lastly, S. pyogenes proteins like scpA15 and SPy_0843, involved in virulence, were also found to interact with DGLA. The modulation of these proteins may prevent the pathogen from evading the host immune response, rendering it more vulnerable to immune defenses (39).
These insights underscore the therapeutic potential of DGLA as an adjunct treatment for bacterial infections. Future research should focus on in vivo studies to validate these interactions and explore the efficacy of DGLA in clinical settings. By targeting specific virulence factors, DGLA could serve as a powerful tool in the fight against antibiotic-resistant bacteria.
Virulence Factor Prediction
Every pathogen, whether it’s fungi, protozoa, viruses, or bacteria, generates virulence factors that allow them to cause infections and harm the host. These virulent factors can be predicted using bioinformatics tools such as VICMpred and VirulentPred, which rely on PSI-Blast and the support vector machine (SVM) method to predict virulent protein sequences (18, 20).
Predictions like these can help in selecting interesting vaccine/drug targets. In this study, VICMpred and VirulentPred tools were employed to analyze 1 protein (epiP) from S. aureus, 10 (PA5371, tesB, PA2871, PA5519, PA3829, PA3741, PA3695, PA2693, PA2218, and PA2168) from P. aeruginosa, and 3 (scpA15, SPy_0843, and cepA) from S. pyogenes. epiP Protein from S. aureus were found to be virulent by both the software. SPy_0843, and cepA from S. pyogenes, also found to be virulent by both the software. Rest of the HPs were predicted to be involved in either metabolic or cellular processes.
Some proteins of P. aeruginosa include HPs, that is, PA5371, tesB, PA3829, PA3741, PA2168, were found to be virulent by VirulentPred. Detailed results of this prediction are listed in Table 2.
Staphylococcus aureus, a Gram-positive bacterium that forms clusters, is commonly found on human and animal skin as well as in the respiratory tract. It is a significant cause of nosocomial infections, particularly methicillin-resistant S. aureus (MRSA) strains. MRSA poses a major public health risk in hospitals, prisons, and nursing homes, especially for individuals with open wounds, invasive devices like catheters, and weakened immune systems. Staphylococcus aureus efficiently colonizes biomaterials used in medical implants, and in cases of biomaterial-associated infections, device replacement becomes necessary, burdening patients while the risk of relapsing infections persists (40). Staphylococcus aureus that using in this study is S. aureus COL. The S. aureus isolate COL, a representative member of this early MRSA lineage first identified in the 1960s (41). Staphylococcus aureus COL have 1 protein (epiP) that interacts with DGLA.
Ribosomally synthesized and post-translationally modified peptides (RiPPs) are an ever-expanding group of peptidic natural products with diverse chemical structures and biological activities. They are mainly isolated from microbes, plants, and animals (42). Gram-positive bacteria synthesize ribosomally synthesized and post-translationally modified peptides (RiPPs) with antimicrobial properties, known as lantibiotics. These lantibiotics serve as potent antibacterial agents against other gram-positive bacteria (43). Proteins work in a cascade to modify lantibiotics post-translationally into their mature form and shield the producing organism from their effects using immunity proteins. In fact, bacteria that produce lantibiotics safeguard themselves by expressing immunity proteins, and these protein-coding genes are typically clustered on plasmids or chromosomes (5). Typically, lantibiotic gene clusters contain a gene encoding a serine protease responsible for processing the leader peptide of the lantibiotic precursor. For instance, the serine protease epiP is involved in the processing of the leader peptide in epidermin biosynthesis (5, 44). The epiP gene contains a peptidase_S8 domain that is present in subtilisin-like serine proteases and is also present in protective antigens of several other species. With DGLA targeting epiP in S. aureus COL, it is very likely to be a candidate for a novel compound against MRSA bacteria.
Pseudomonas aeruginosa, an opportunistic pathogen, causes severe hospital-acquired infections in immunocompromised patients. It also leads to lung infections in cystic fibrosis patients and is common in severe burn wounds. Key factors contributing to its clinical pathogenicity include biofilm formation, virulence factor production, and antibiotic resistance, making treatment increasingly challenging. Consequently, there’s a pressing need for novel antimicrobial agents to combat this significant pathogen (45).
Pseudomonas aeruginosa have 10 protein that interacts with DGLA. 5 proteins include HPs, that is, PA5371, tesB, PA3829, PA3741, PA2168, were found to be virulent by VirulentPred. 3 proteins which belong to the family of acyl-CoA thioesterase are, tesB, PA5371, and PA3741. The two HPs mentioned last, also family of Cytosolic acyl coenzyme A thioester hydrolase, after examine using InterPro tool to Classification of protein families. Acyl-CoA thioesterases (ACOTs) are enzymes that catalyze hydrolysis of fatty acyl-CoAs to free fatty acids and CoA-SH. ACOTs are expressed ubiquitously in prokaryotes and eukaryotes (46). Thioesterase that has relatively broad substrate specificity, hydrolyzing primarily medium- and long-chain acyl-CoA substrates to free fatty acids and CoA. Functions in the thioesterase-dependent pathway of beta-oxidation of oleate and conjugated linoleate ((9Z,11E)-octadecadienoate or CLA), which provides all energy and carbon precursors required for the growth of E.coli.
The Next Virulent HPs Protein are PA3829 and PA2168. Both HPs belong to the family FrsA esterase according to the InterPro. The FrsA family proteins function as esterases, catalyzing the hydrolysis of esters. These proteins play diverse roles in biological processes, including breaking down cutin (a plant cuticle polyester) and synthetic polyesters like poly(ethylene terephthalate) PET. Some family members are involved in complex biosynthetic pathways for natural products, such as mycotoxins and antibiotics. Additionally, certain FrsA proteins disrupt bacterial communication as quorum-quenching agents. Moreover, specific FrsA family proteins contribute to fungal virulence and plant pathogenicity, participating in melanin biosynthesis and producing fungal signals recognized by resistant plants.
The Fermentation-respiration switch protein (FrsA) was initially discovered as a novel protein that specifically binds to the unphosphorylated form of IIAGlc in Escherichia coli extracts. FrsA is believed to play a role in regulating the metabolic balance between respiration and fermentation pathways by interacting with IIAGlc. When the frsA gene in E. coli is disrupted, it leads to an increased respiration rate when glucose serves as the carbon source. Conversely, overexpression of FrsA accelerates the fermentation rate on glucose, resulting in the accumulation of mixed-acid fermentation products. Although FrsA’s physiological function is associated with metabolism, its precise biochemical role remains unclear (47).
Streptococcus pyogenes, a significant human-specific bacterial pathogen, leads to various manifestations, including mild localized infections and life-threatening invasive ones. Inadequate treatment of S. pyogenes infections can result in postinfectious complications like acute rheumatic fever and post-streptococcal glomerulonephritis. Additionally, it causes severe invasive infections such as necrotizing fasciitis and toxic shock syndrome, which are associated with high morbidity and mortality.
Streptococcus pyogenes have 3 protein that interacts with DGLA, all 3 protein are virulent according to VirulentPred. The first protein is scpA15 which is family of C5A peptidase. Most pathogenic Streptococci, with the exception of S. pneumoniae, produce a potent enzyme called C5a peptidase. This enzyme, which is highly conserved and found on the bacterial surface, plays a crucial role in evading the human immune system by inactivating complement proteins (C3, C5a, and C3b). Additionally, it acts as an adhesion factor during host invasion by binding to integrin, fibronectin, and epithelial cells. Despite decades of research, attempts to develop a vaccine targeting C5a peptidase have been unsuccessful, and there are no reports of small molecule inhibitors for this enzyme (48)
The second virulent protein of S. pyogenes that interacts with DGLA is SPy_0843. After examine using InterPro tool to Classification of protein families, SPy_0843 is categorized belong to Putative surface bspA-like protein family. These proteins are associated with interactions involving host tissues. Within this protein family, individual members may participate in adherence or invasion processes during the infection cycle, potentially by binding to host cell surfaces or components of the extracellular matrix. However, the precise molecular mechanisms and the specific host tissues targeted by these proteins have yet to be fully understood. Gaining insight into the function of these proteins could offer valuable information about pathogenic strategies used by microbes and may have implications for developing therapeutic interventions. Research has demonstrated that BspA-like proteins play a role in bacterial adherence, epithelial cell invasion, and binding to fibronectin and fibrinogen. A study provided direct evidence of BspA’s in vivo contribution to pathogenesis by showing that BspA-deficient bacteria were notably less pathogenic than their wild-type counterparts (49).
The third virulent protein of S. pyogenes that interacts with DGLA is cepA which is family of cell envelope proteinase. Among the various proteins, cell-envelope proteinases (CEPs) have garnered significant attention and are extensively utilized for biotechnological purposes. These CEPs play crucial roles in the proteolytic system such as in lactic acid bacteria (LAB), as they are essential for breaking down proteins in the growth media into peptides and amino acids needed for LAB’s nitrogen nutrition (50).
B-cell Epitope Analysis & Subcellular Location Analysis
We carry out analysis the B-cell epitope locations of the virulent proteins from S. aureus, P. aeruginosa, and S. pyogenes using BepiPred v2.0. BepiPred-2.0, a web server for predicting B-cell epitopes from antigen sequences, outperforms other available tools in sequence-based epitope prediction. It achieves high accuracy on test datasets and validation datasets, providing specific locations on bacterial proteins that can bind to antibodies or the immune system, as identified through bioinformatics (22). Antibodies play a vital role in the immune system of vertebrates. They recognize and counteract foreign substances like viruses, bacteria, fungi, cancer cells, and toxins by binding to specific regions on their surface, known as antigens. These specific antibody parts, called epitopes, are the focus of B-cell epitope analysis, aiming to identify robust epitopes for potential use in diagnostics and epitope-based vaccines (51).
The analysis findings indicated that certain amino acid sequences in each protein can bind to B cells (referred to as epitopes). Additionally, the study clarified that molecular interactions occur between proteins from S. aureus, P. aeruginosa, and S. pyogenes, along with DGLA. These interactions are manifested as B-cell binding, visually represented by yellow epitopes (Figure 2). Antibacterial compounds, such as DGLA, binding to specific locations activate an immune response that generates antibodies. Additionally, DGLA’s attachment to epitopes prevents pathogenic bacteria from adhering to the host cell membrane, where it disrupts bacterial peptidoglycan structure.
Based on the psortb examination, researchers identified various subcellular locations for functional proteins that interact with DGLA (Table 4). The proper subcellular localization of proteins is crucial because it determines their access to interacting partners, post-translational modification machinery, and integration into functional biological networks. Aberrant protein localization has been associated with drug targets developing (52).
For example in S. aureus COL have epidermin leader peptide processing serine protease (epiP) as functional proteins. epiP, a subtilisin-like extracellular epidermin leader peptidase, plays a role in processing the mature lantibiotic epidermin such as in Staphylococcus epidermidis. EpiP was anticipated to be located extracellular localization due to the existence of a leader peptide and the absence of other recognized signals for anchoring to the membrane or cell wall (5).
Future Directions for Validation
To enhance the robustness of our findings, it is crucial to pursue empirical validation of the bioinformatics predictions presented in this study. Several experimental approaches can be employed to confirm the interactions and virulence factors associated with DGLA:
- In Vitro Binding Assays: Conduct binding assays such as surface plasmon resonance (SPR) or isothermal titration calorimetry (ITC) to directly measure the interaction affinities between DGLA and the identified virulent proteins from S. pyogenes, S. aureus, and P. aeruginosa.
- Mutagenesis Studies: Generate site-directed mutants of the virulent proteins, particularly those identified as potential DGLA interaction sites, to assess the impact on their function and binding capabilities. This approach will help pinpoint critical residues involved in the interactions.
- Protein Localization Studies: Utilize fluorescent tagging and microscopy techniques to observe the subcellular localization of virulent proteins in the presence and absence of DGLA. This will provide insights into how DGLA influences the spatial distribution of these proteins within the bacterial cells.
- In Vivo Infection Models: Implement animal models of infection to examine the effects of DGLA on bacterial virulence and pathogenicity. Monitoring the progression of infections with and without DGLA treatment will offer valuable data on its therapeutic potential.
- Transcriptomic and Proteomic Analyses: Perform transcriptomic and proteomic analyses to investigate the broader impact of DGLA on bacterial gene expression and protein production. This will help elucidate the pathways and processes affected by DGLA interaction.
Potential Limitations
Despite the significant findings of this study, it is essential to acknowledge certain limitations, particularly the reliance on computational predictions and bioinformatics tools without empirical validation. While these methods provide valuable insights, they may not fully capture the complexity of biological systems, and the predicted interactions require experimental confirmation to validate their accuracy. The absence of in vitro or in vivo studies to support the computational data could impact the interpretation of results, and future research should focus on empirical validation to strengthen the conclusions drawn from this study.
This study proves that Dihomo-γ-linolenic acid (DGLA) as a natural chemical compound isolated and identified from Streptomyces actinomycinicus PJ85 effectively acts as an antibacterial agent, affecting virulence factors by interacting with several proteins of S. aureus, P. aeruginosa, and S. pyogenes through bioinformatics. It is known that virulent proteins in Staphylococcus epidermidis and S. pyogenes interacting with DGLA are located in the Extracellular, cytoplasmic, and cellwall indicating that DGLA's ability to bond with drug targets.
Special thanks to our colleagues in Palangka Raya University for their invaluable feedback and discussions that greatly improved the manuscript.
Ethical Considerations
The researcher confirms that this study was conducted in full compliance with ethical standards. No ethical violations occurred during the research process.
Authors’ Contributions
Nawan Nawan: Conceptualization, Methodology, Writing - Original Draft, Supervision, Software, Validation, Project Administration. Septi Handayani: Data Curation, Formal Analysis, Writing - Review & Editing. Agnes Immanuela Toemon: Investigation, Resources, Visualization. All authors have read and approved the final version of the manuscript. The final manuscript was read and approved by all of the authors.
Conflicts of Interest
This research project was conducted without any financial support from external sources.
The authors were not utilized AI Tools.
Received: 2024/12/12 | Accepted: 2025/03/13 | ePublished: 2025/03/30
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |