BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-2454-en.html
2- Department of Biology, College of Science, University of Kirkuk, Kirkuk, Iraq ,
3- Department of Chemistry, College of Science, University of Kirkuk, Kirkuk, Iraq
Scopulariopsis brevicaulis (S. brevicaulis) is a saprophytic fungus commonly found in soil, vegetables, air, and decaying organic waste (1). To date, there are eight species of Scopulariopsis that cause human infections. Of these, Scopulariopsis brevicaulis is non- dermatophyte filamentous fungus that rarely causes infections in humans (2, 3). However, recent studies reported an increase in the number of infections caused by this fungus (4). S. brevicaulis causes onychomycosis with an estimated rate of 3-10% (5). The most important clinical manifestation of onychomycosis is distal and lateral subungual onychomycosis (DLSO) (6, 7). S. brevicaulis infections can occur in the smooth skin and subcutaneous tissues. Disseminated S. brevicaulis infections like endocarditis, sinusitis, endophthalmitis, pneumonia, and brain abscess have also been reported (5, 8). These types of infections are difficult to treat and cause high fatality rate particularly in immunocompromised patients (5, 9). Several predisposing risk factors like familial dermatoses, trauma, diabetes mellitus, and peripheral circulatory insufficiency are commonly associated with S. brevicaulis infections (4).
Recent studies have also reported inefficiency of commonly used antifungal agents like amphotericin B (AMB) against S. brevicaulis, which represents a major concern (10). Molecular tools and genomic sequencing are commonly used for the identification of fungi. Recent studies have highlighted the genetic variability among S. brevicaulis isolates from different clinical sources, necessitating molecular approaches for the accurate identification and phylogenetic analysis (11).
Several genomic regions, including D1/D2, EF-1α, and ITS have been successfully utilized to differentiate the species among Scopulariopsis and related genera. Molecular techniques provide faster, more sensitive, and more accurate identification of dermatophytes compared to the culture methods (3). Traditional methods are time-consuming and occasionally produce unreliable results. Additionally, relatively late development of characteristic brown-powder configuration of S. brevicaulis colonies presents a further diagnostic challenge (12, 13).
Internal transcribed spacer regions (ITS1 and ITS2) can be used as molecular targets for the identification of intra-species variation in S. brevicaulis and for the phylogenetic analysis. This genetic region, situated between the small (18S) and large subunits (28S) of rRNA genes in the fungal genome, is known for its variability among different fungal strains and species. By comparing these sequences with reference databases such as GenBank, researchers can confirm the identity of S. brevicaulis isolated from the clinical or environmental samples (8).
A thorough taxonomic molecular study related to S. brevicaulis is still lacking in Kirkuk city, Iraq. In order to improve our understanding about diversity and genetic capabilities of this fungus we endeavoured to isolate S. brevicaulis from the skin lesions and identify to molecular level. This would assist in defining the species spectrum and relative frequencies of Scopulariopsis in clinical samples. Phylogenetic tree was then characterized based on the ITS sequence analysis. Finally, the in vitro antifungal sensitivity test was also determined using disc diffusion method.
Fifty nail and skin samples were collected from diabetic patients suffering from skin infections at dermatology clinics in Kirkuk hospitals (Kirkuk, Iraq) during the period from April 2023 to November 2023. The patients' ages ranged from 15 to 60 years, and both sexes were included. All selected patients were confirmed to have diabetes. The Collected samples were initially subjected to direct microscopic examination using potassium hydroxide (KOH). Briefly, a piece of the sample was placed on a clean slide and 1-2 drops of 10% KOH was added. The prepared slide was then examined under a light microscope (40X). All fungal isolates were further characterized by culturing on Sabouraud dextrose agar (SDA) plates and incubating for two weeks at room temperature (5). Chromogenic Candida agar (CCA), a differential media was also used for identification and differentiation of Candida species (14).
Microscopic properties of isolated fungi (shape, size, and arrangements) were also studied. A loopful colony mixed with lactophenol methylene blue was examined under light microscope (40X) (15, 16). Biochemical tests such as urease, hair perforation and germ tube tests were also used for identification of fungal species (17, 18). On the other hand, bacterial isolates were identified based on the culture, microscopic, and biochemical tests (19).
DNA Extraction
Fungal DNA was extracted from S. brevicaulis using Fungal DNA MiniPrep™ kit following the manufacturer’s protocol. Fungal cells grown on Sabouraud dextrose agar were resuspended with phosphate buffer saline into ZR Bashing Bead™ Lysis Tube. Lysis buffer was added and left in a bead beater for at least 5 min. The tube was then centrifuged at 10000 xg for 1 min. The supernatant was transferred to the Zymo-Spin™ IV Spin Filter in a collection tube and centrifuged at 7000 xg for 1 min. DNA Binding buffer was added to the filtrate in the collection tube. The mixture was transferred to the Zymo-Spin™ IIC Column in a collection tube and centrifuged at 10000 xg for 1 min. DNA Pre-Wash Buffer was added to Zymo-Spin™ IIC Column in a new collection tube. The washing and centrifugation steps were repeated. The extracted fungal DNA was eluted in Elution Buffer and kept at −20°C for further use.
Molecular Detection
The extracted DNA was amplified using an automated PCR thermal cycler (Bioneer, Korea). The primers against ribosomal subunit conserved region of the large subunit rRNA (ITS 1) are listed in Table 1. The primers were designed based on the Mushıb et al. study (Macrogen, Korea) (20). The amplification reaction was achieved in a 25 μL reaction mixture containing primers (10 pmol/each), DNA template, Taq PCR PreMix (Intron, Korea), and distilled water. The PCR program was set for 35 cycles after an initial denaturation at 94°C for 3 min. Each cycle included three steps: denaturation at 94°C for 45 sec, followed by annealing at 58°C for 1 min, and an extension at 72°C for 45 sec. The final extension step was set to 72°C for 7 min. The PCR amplicon was then examined electrophoretically on 1.5% agarose gel stained with ethidium bromide for 45 min at 100 volts and visualised under UV light. The Amplified product was determined by comparison with the standard DNA ladder.
Table 1. The primers specifications
| Primer | Sequences 5’-3’ | Product size (bp) |
| Forward | 5`-TCCGTAGGTGAACCTGCGG-3` | 550 |
| Reverse | 5`-TCCTCCGCTTATTGATATGC-3` |
The amplified products underwent purification and stored at -20°C until sequencing. DNA sequencing was conducted bi-directionally using the same primer set employed for amplification (Table 1) using Sanger analysis software.
Phylogenetic Tree Analysis
The raw sequences data was checked for the quality in MEGA software. The obtained sequences were trimmed by BioEdit software (21) and put into G-blocks 0.91b in order to obtain good quality sequences, and to eliminate the ambiguously aligned positions and divergent regions prior to phylogenetic analysis (22). The resulted sequences were aligned with those available for S. brevicaulis in the GenBank, including 14 in-group taxa using MAFFT 7.520 software.
Multiple sequence alignments were performed using MEGA software version 11.0.13 with Clustal W application. They were manually refined within the same software platform. Nucleotide sequences of ITS from the clinical isolate and 14 taxa retrieved from NCBI GenBank were used for the construction of phylogenetic tree using maximum likelihood (ML) and Bayesian inference (BI) analysis under MEGE version 5.05. Mr Bayes v. 3.2.6 was used to detect BI analyses with run by partition codon positions (23).
Markov-chain Monte-Carlo searches version 3.2.1 involved four chains, each running for 10,000 generations. Tree was sampled every 10 generations during each chain run. The initial 25% of tree from each run was excluded as burn-in, and the remaining trees were combined to form a single tree using 50% majority rule consensus approach. Bayesian inference posterior probability (BI-PP) values equal to or above 0.95 were found to be statistically significant. Furthermore, evolutionary relationships and divergence points among different isolates were studied using evolutionary distances. This would assist to elucidate how the examined strains are interconnected through their genetic history.
Nucleotide Sequence Deposition in GenBank
The fungal isolate from the current study was deposited in GenBank under the accession number OQ236576.1.
Antifungal Susceptibility Testing
Disc diffusion method was used to detect the susceptibility of the isolated fungus against five antifungal agents (amphotericin B (AMB) (20µg), Ketoconazole KCA (10 µg), Clotrimazole CLO (50 µg), Fluconazole FLU (25 µg) and Nystatin NY (100 µg) (Liofilchem, Italy) (24). This test was performed in accordance with NCCLS reference method (M51-A) (25).
S. brevicaulosis was initially inoculated on Sabouraud dextrose agar (supplemented with chloramphenicol and cyclohexadiene), and incubated for 7 days at 25℃. After incubation, the fungal suspension was prepared from fresh culture by mixing fungal colonies with 3 ml of sterile distilled water. The inoculum was then transferred to the SDA plates with a sterile cotton swab and left to dry. Using sterile force, antifungal discs mentioned above were placed into inoculated plate and incubated for 5-10 days at 28℃. Following incubation, the results were interpreted and the inhibition zone around discs was measured in mm using ruler.
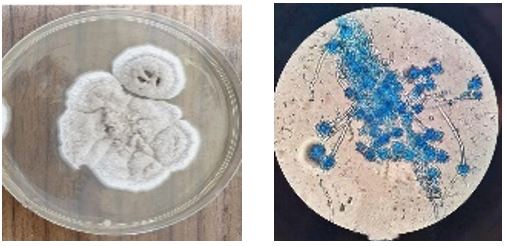
Interestingly, a single strain of S. brevicaulis (4%) was isolated from a diabetic patient. This isolate appeared as flat, velvety, or powdery, light brown colonies when cultured on Sabouraud dextrose agar. Under microscope, conidia formed dark brown chains originating from a conidiogenous cell known as an annellide, as illustrated in Figure 1.
Table 2. Fungal and bacterial isolates recovered from nail and skin samples.
| Clinical samples | |||||||
| Fungal isolates (25) | Bacterial isolates (5) | ||||||
| Mold | No. (%) | Yeast | No. (%) |
Gram negative | No. (%) |
Gram positive |
No. (%) |
| Scopulariopsis brevicaulis | 1 (4%) |
Candida albicans | 7 (28%) |
Pseudomonas aeruginosa | 2 (40%) |
--- | --- |
| Trichophyton mentagrophytes | 14 (56%) | Candida glabrata | 3 (12%) |
Klebsiella spp | 1 (20%) |
Staphylococcus aureus | 2 (40%) |
A B
Figure 1. (A)- Scopulariopsis brevicaulis on Sabouraud dextrose agar. (B)- S. brevicaulis stained with lactophenol dye examined under light microscope (40X).
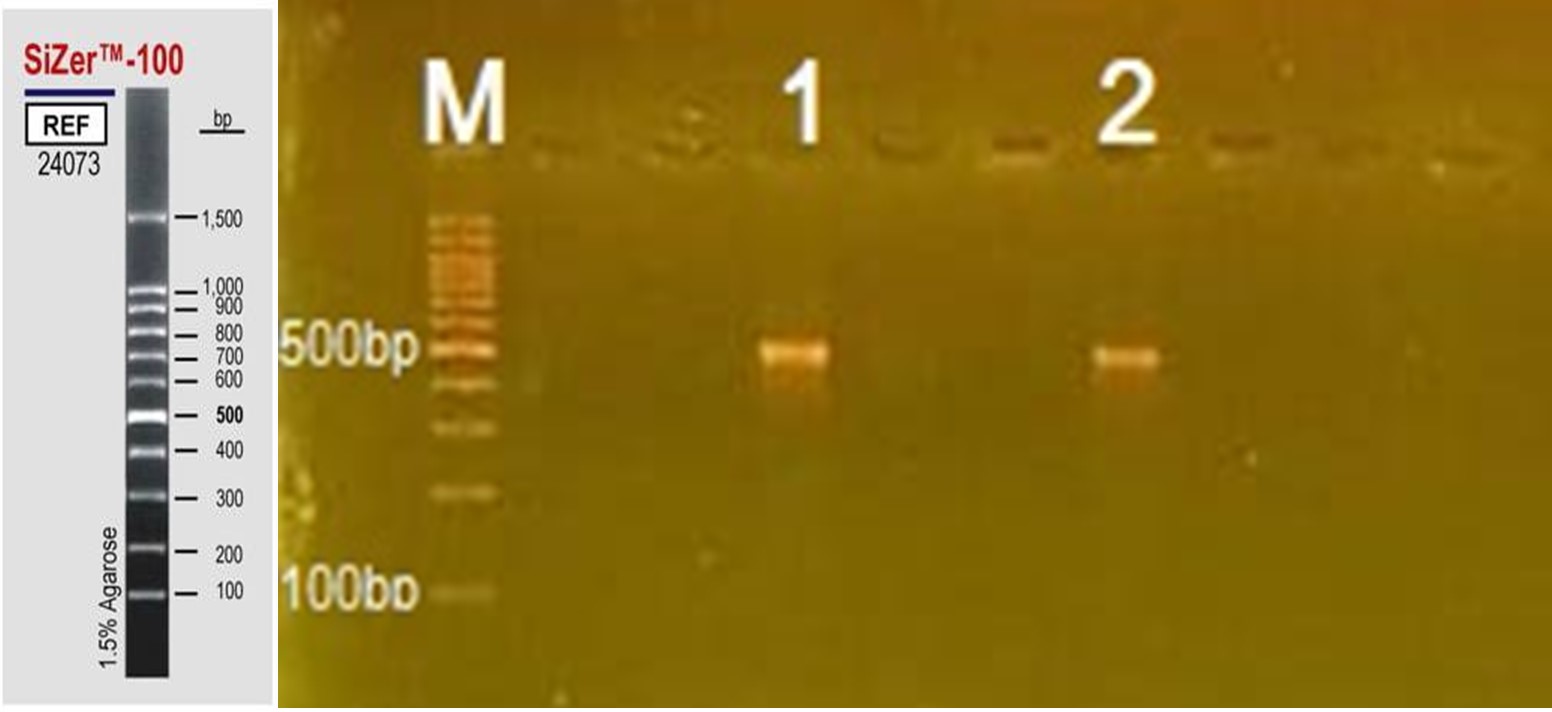
A PCR-based method was used for the specific detection of S. brevicaulis using ITS1 region. The results showed successful amplification of 550 bp DNA fragment corresponding to the ribosomal large subunit RNA gene of Scopulariopsis as shown in Figure 2.
In addition, nucleotide variation between the submitted sequences and those deposited in GenBank ranged from 2 to 6, which indicate specific genetic variation between submitted and reference sequences. For example, sequence with accession number KX923976.1, submitted from the Netherlands in 2017, has 98.96% similarity and 98% query coverage with 6 nucleotide differences. Similarly, sequence data OL589623.1, submitted from South Korea in 2022, shows 98.98% similarity and 99% query coverage, also with 6 nucleotide differences. These details provide insights into the genetic variation and geographic distribution of the analyzed sequences.
As shown in Figure 3, the clades are grouped in three main clusters based on the degree of evolutionary relationships. S. brevicaulis clinical isolate clade was grouped together with those isolated from the United States of America (USA). However, this clade was far away from those recovered from Iraq (Basrah) and neighbouring countries (Turkey and Iran).
The evolutionary distances represent the genetic divergence between the clinical sample (n2023_sample) and various other taxa arranged from the highest to the lowest (Table 3).
Table 3. Sequences data of S. brevicaulis strains retrieved from GenBank
| Accession number | Submission date | Similarity (%) | Query coverage (%) | Number of Nucleotide differences | Evolutionary distances | Country of submission |
| KX923976.1 | 2017 | 98.96 | 98 | 6 | 0.006743741 | Netherlands |
| OL589623.1 | 2022 | 98.98 | 99 | 6 | 0.01056835 | South Korea |
| OP752128.1 | 2022 | 98.63 | 99 | 5 | 0.0042839377 | Turkey |
| LC638842.1 | 2021 | 98.63 | 99 | 5 | 0.006508003 | Basrah Iraq |
| OW982815.1 | 2022 | 99.14 | 99 | 4 | 0.0042428491 | Belgium |
| KP641165.1 | 2015 | 99.13 | 98 | 3 | 0.004022939 | Iran |
| EU821474.1 | 2015 | 99.48 | 98 | 3 | 0.0043055723 | Colombia |
| MT576462.1 | 2020 | 99.48 | 98 | 3 | 0.0042817352 | China |
| AY625065.1 | 2016 | 99.48 | 99 | 3 | 0.000547002 | USA |
| KP132734.1 | 2015 | 99.49 | 99 | 3 | 0.012336815 | Australia |
| MT316372.1 | 2020 | 99.49 | 99 | 3 | 0.0008799436 | Italy |
| OR366529.1 | 2023 | 99.14 | 99 | 2 | 0.003763237 | India |
| MF156019.1 | 2019 | 99.31 | 99 | 2 | 0.000723214 | Czech Republic |
| AY773332.1 | 2004 | 99.65 | 98 | 2 | 0.002655759 | Australia |
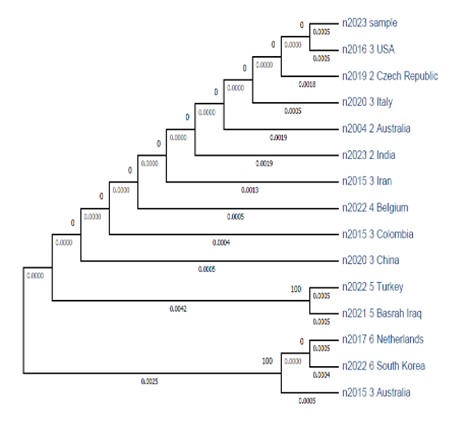
Figure 3. Phylogenetic tree of S. brevicaulis. This tree was constructed using maximum likelihood (ML) and Bayesian inference (BI) analysis based on the ITS sequences with MEGE version 5.05. The ITS sequences of our isolate (labeled as n2023 sample) were aligned with 14 S. brevicaulis sequences retrieved from GenBank using MAFFT 7.520. Multiple sequence alignments were carried out using MEGA software version 11.0.13 with the Clustal W application.
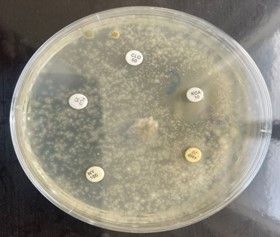
Figure 4. Antifungal sensitivity test of Scopulariopsis brevicaulis.
Table 4. Sensitivity and resistance profile of antifungal agents against Scopulariopsis brevicaulis
| Antifungal agents | Inhibition zone diameter (mm) |
| Ketoconazole (KCA) | 15 mm |
| Clotrimazole (CLO) | 0 |
| Fluconazole (FLU) | 0 |
| Amphotericin B (AMB) | 0 |
| Nystatin (NY) | 0 |
Scopulariopsis brevicaulis is known to cause a variety of infections in humans, ranging from superficial skin infections to potentially life-threatening invasive mycoses (28). Despite its clinical significance, data on the prevalence of S. brevicaulis in skin infections in Kirkuk city, Iraq is limited. Thus, we attempted to isolate and identify S. brevicaulis from nail and skin infections. S. brevicaulis was recovered from 4% (n=1) of our fungal positive clinical samples. This lower prevalence rate of S. brevicaulis among fungal infections aligns with other literature data. A study done by Petanović et al (4) reported that only 2.2% of the fungal isolates was S. brevicaulis. In a study conducted by Azar et al (29), a lower prevalence rate of S. brevicaulis was observed with only 3.30% of diabetic patients being affected. Similarly, the prevalence of S. brevicaulis was also low in Korea with low reported rates (1.23% and 1.41%) (30). Furthermore, Turkish researchers demonstrated that only 3% of onychomycosis was caused by S. brevicaulis (20). Dhib et al. (31) also reported that only 0.3% of fungal positive nail cases were S. brevicaulis. Additionally, Bassiri-Jahromi and Khaksar demonstrated that S. brevicaulis is responsible for 2.1% of non-dermatophytic onychomycoses cases (32). Consistent with our results, Tosti et al. found S. brevicaulis in 3.9% of positive cases (33). Similarly, S. brevicaulis was identified in only 3.5% (28). On the other hand, some studies indicate higher prevalence rates of S. brevicaulis in onychomycoses, at 42.8% (34) and 43.5% (35). Variation of prevalence rate might be attributed to the geographic differences in mold distribution, variations in diagnostic criteria of onychomycosis, and the use of different culture methods (30). Other fungal isolates were also identified. Trichophyton spp. exhibited the highest prevalence rate (56%) followed by Candida spp. that accounted for 40%. These results agree with the findings of Mushıb et al (20) and Naik et al (36).
The ITS 1 region of our isolate was successfully amplified using PCR, similar to those reported in other researches (20). By sequencing the ITS region of S. brevicaulis, researchers can identify variations in the nucleotide sequences distinguishing different strains within the species (27). These variations provide insights into the genetic diversity, population structures, epidemiology, pathogenicity, and response to the antifungal treatments as well as evolutionary relationships among different isolates of S. brevicaulis.
To the best of our knowledge, this is the first study employing ITS based on phylogeny to determine the genetic variation in S. brevicaulis. Our phylogenetic tree revealed that the isolated fungus belongs to the same clade as those reported in the USA (accession number AY625065.1). However, it differs from the isolates found in the same country Iraq (Basrah), as well as in neighbouring countries like Iran and Turkey. This suggests that there are evolutionary distances between the isolates within the same region, which might contribute to the development of antifungal resistance.
Thus, focusing on intra-species variation of S. brevicaulis in a single country enables the researchers to identify distinct genetic variants or clades within S. brevicaulis populations across different regions or environments within the country. Furthermore, it assists in understanding which ITS variants are more prevalent in specific regions or environments within the country providing insights into the environmental factors affecting fungal distribution. Our findings agree with the previous data (37) showing the resistance of S. brevicaulis to the most antifungal agents particularly amphotericin B and fluconazole, except for ketoconazole, which displayed an inhibition zone of 13.4 mm. These drugs are the most important antifungals commonly used for the prophylaxis and treatment of mold infections. Numerous studies on in vitro antifungal activity and their combinations against clinical isolates of S. brevicaulis have confirmed it as a multi-drug resistant pathogen (38-40). Even with advancements in antifungal treatments, managing onychomycosis is still difficult, with over 25% of the patients showing incomplete or no response to treatment (30, 41, 42).
Multi-drug resistance pattern observed on the current and previous studies highlights the challenging in the treatment and management of infections caused by S. brevicaulis. More researches are required for understanding the resistance mechanisms, and exploring alternative treatment options to improve the patient outcomes and address the challenges posed by this pathogen.
Despite these findings, our study has some limitations. Firstly, it was based on a single isolate of S. brevicaulis which restricts the generalizability of our findings and highlights the need for more extensive sampling. Secondly, the study was confined to the isolates from Kirkuk city. Expanding the geographic scope would offer a more comprehensive understanding of the evolutionary relationships and distribution patterns of S. brevicaulis.
This study highlights the importance of S. brevicaulis identification in diabetic patients due to its multi-drug resistant property against antifungal drugs. Accurate identification of S. brevicaulis is vital for selecting the appropriate treatments to prevent severe mycoses. Furthermore, DNA sequencing and phylogenetic analysis showed genetic divergence from the strains reported in other countries and moderate evolutionary distance from those found in the same country (Basrah, Iraq). Thus, large-scale clinical trials using advanced molecular techniques are needed to monitor the evolutionary changes and resistance profiles of S. brevicaulis, guiding the development of effective treatments.
The authors express their gratitude to Department of Biology, College of Science, University of Kirkuk, for their assistance and support on this project.
Ethical Considerations
University of Kirkuk, Science College, Biology Department Ethics Committee approved the study under the code number ScB20.
Authors’ Contributions
All authors contributed equally to the preparation of this research article including, study concept and design, data collection, analysis and interpretation, and drafting and revision of the manuscript.
The author(s) received no financial support for the research or publication of this article.
Conflicts of Interest
Received: 2024/07/7 | Accepted: 2024/09/4 | ePublished: 2024/09/29
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |