BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-2351-en.html
2- Department of Bacteriology, Faculty of Medical Sciences, Tarbiat Modares University, Tehran, Iran ,
3- Department of Biostatistics, Faculty of Medical Sciences, Tarbiat Modares University, Tehran, Iran
4- Division of Colon and Rectal Surgery, Department of Surgery, Imam Khomeini Hospital Complex, Tehran University of Medical Sciences, Tehran, Iran
5- Infectious Diseases and Tropical Medicine Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Clostridioides difficile (C. difficile) stands as the primary cause of colitis, toxic megacolon, antibiotic-associated diarrhea (AAD), and pseudomembranous colitis (1, 2). Over the last twenty years, an escalation has been observed in both the severity and frequency of C. difficile infection (CDI). Notably, almost 30% of individuals with antibiotic-associated diarrhea (AAD) have been verified to suffer from CDI (3).
The colorectal cancer (CRC) is ranked as the third most prevalent cancer worldwide in terms of morbidity. The primary symptoms often encompass weight loss, abdominal pain, bleeding, anemia, and alterations in the bowel habits. It is noteworthy that a significant proportion of the CRC cases occur in individuals with no family history of the disease (4, 5). While advanced age is recognized as a risk factor for the CRC, there is a growing trend of the disease incidence among younger individuals (6). Despite a well-established understanding of the CRC pathogenesis, recent studies have emphasized the significant influence of the gut microbiota in the development of this cancer. Notably, antibiotic treatment has the potential to disrupt the normal balance of the gastrointestinal microbiome, creating a favorable environment for the growth of various pathogens in the luminal region of the intestine (7).
Numerous investigations have highlighted the significant role of certain bacterial pathogens in the CRC deterioration. Specifically, research focused on CDI has revealed its association with increased morbidity and mortality rates (8). Additionally, CDI has been linked to the increased risk of hospitalization, discontinuation of post-surgical complementary treatment, and elevated costs in the CRC patients. Recent reports have indicated that 17% of the CRC patients are affected by CDI. Additionally, factors like the use of antibiotics, colonic involvement, and chemotherapy have been identified as significant risk factors associated with the onset of CDI in the CRC patients (9). Most of the gut microbiota dwell in areas near the intestinal epithelial surface. A variety of bacteria in the intestine significantly contribute to the early-life development of the immune system while also safeguarding against colonization by pathogens (8). Nonetheless, infection or colonization of the intestine by the pathogens or pathobionts, along with the metabolites they release, could modify the microbiota population in the gastrointestinal tract (10). Limited data are available regarding fecal carriage and intestinal colonization of C. difficile in the CRC patients. Thus, this study aimed to investigate the presence of C. difficile in the patients with colorectal cancer compared to the healthy individuals in Iran.
Definition of Patients and Healthy Volunteers
The CRC patients included in this study were people who referred to the Colorectal Department of a major Hospital in Tehran, Iran, from June 2022 to May 2023. Patients with symptoms such as constipation, iron deficiency anemia, diarrhea, or rectal bleeding were subjected to confirmatory tests for the CRC diagnosis. Colorectal cancer was confirmed by colonoscopy and pathological examination. The confirmed CRC patients were subjected to a cancer staging process with computed tomography (CT) of the chest and the entire abdominal and pelvis areas, magnetic resonance imaging (MRI), ultrasonography of the middle part of the body, and then a blood test to check CEA (11) according to the tumor, node, metastasis (TNM) staging system provided by the American Joint Committee on Cancer (AJCC) (12, 13).
In this study, 25 CRC patients along with 25 healthy individuals as the control group were included. The patient and healthy controls were meticulously matched with respect to the ethnicity, sex, and age for the optimal comparability in the study. The two study groups, CRC patients and healthy individuals, were selected from people with no underlying diseases. The subjects with any of the following criteria were excluded from the study: presence of tumors or gastrointestinal malignancies (for patients), body mass index (BMI)>30 kg/m2, use of probiotics or antibiotics in the past three months, adherence to a vegetarian diet, chronic bowel disorders and food allergies or any dietary restrictions, and recent surgeries or therapeutic interventions such as chemotherapy or radiotherapy (14, 15). The stool samples were collected from the patients and healthy volunteers. The samples were collected in special stool collection containers and transferred quickly (within two hours after sampling and under cold-chain conditions) to the Bacteriology Laboratory of the Tarbiat Modares University.
DNA Extraction
Total DNA was extracted from 500 mg of the stool samples utilizing a stool DNA isolation kit (Favorgen Biotech, Taiwan) in accordance with the manufacturer's protocol. Gel electrophoresis technique was employed to evaluate the quality of the extracted DNA utilizing 1% agarose gel. Subsequently, DNA quantification was carried out using a nanodrop spectrophotometer (BioTek Epoch, US). The extracted DNA samples were then stored at -20°C, while the remaining stool portions were divided into three containers and stored at -80 °C. The C. difficile ATCC 10898 was used as a control (16).
TaqMan Real-time PCR Assay
To determine the presence of C. difficile in the patient and control groups, the extracted DNA from each sample was assessed through TaqMan real-time PCR targeting the 16S rRNA gene of C. difficile using specific primers and probe. The specific primers and probe related to C. difficile were used to amplify 155 bp amplicons of the 16S rRNA gene (forward primer: 5′-GCAAGTTGAGCGATTTACTTCGGT-3′/ reverse primer: 5′-GTACTGGCTCACCTTTGATATTYAAGAG-3′/ prob: FAM-TGCCTCTCAAATATATTATCCCGTATTAG-TAMRA) (17). These primers were tested by SILVA high-quality ribosomal RNA database (https://www.arb-silva.de) and Primer-BLAST web tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast) for inclusivity, specificity, and probable mismatches. The primers were further analyzed by OligoAnalyzer tool (https://www.idtdna.com/pages/tools/oligoanalyzer) for melting temperature and secondary structures. The annealing temperature was considered a few degrees higher than the melting temperature to overcome the non-specific off-target amplifications. TaqMan real-time PCR was performed by the Stratagene Mx3000p system (Agilent, US). The amplification program consisted of 40 cycles, each cycle comprising denaturation at 94°C for 40 sec and annealing at 60°C for 40 sec. The initial cycle involved a denaturation step at 94°C for 5 min. The DNA of C. difficile strain ATCC 10898 was employed as a standard control. Reactions were conducted in a total volume of 20 μL, including 2 µL of template DNA, 10 µL of 2X SYBR Green real-time PCR master mix (Ampliqon, Denmark), 1 µL of forward primer, 1 µL of reverse primer, and 6 µL of sterilized ultra-pure water (8, 18). A control reaction containing all the reaction components except the template DNA was run in parallel with each amplification reaction as a negative control. Finally, the amplification plots, cycle threshold (Ct) values, and melting curves of the samples were recorded. Each reaction was performed in triplicate, and the mean of triplicate reactions were considered as Ct values.
Statistical Analysis
Data were analyzed using SPSS software Ver. 26.0. Chi-square test was used to investigate the difference between the two patient and healthy groups in terms of the presence of C. difficile in their stool samples. Outcomes exhibiting a P-value<0.05 were deemed to be statistically significant.
Patients and Controls
All 25 patients were in stage III of the disease. The two groups were matched in terms of age and gender with no significant difference. The age range of the studied subjects was between 28 and 68 years. The subjects’ BMI was below 30 kg/m2 (between 22.2 and 29.2 kg/m2). The tumor location was recorded in the colon for all patients in the study. No significant difference was observed between the two groups with regard to smoking and alcohol consumption (P>0.05). The robustness of the results to the sample size was confirmed by consulting the guidelines of Tabachnick and colleagues (2013) (19).
Inter-group Real-time PCR Analysis of C. difficile Between CRC and Healthy Groups
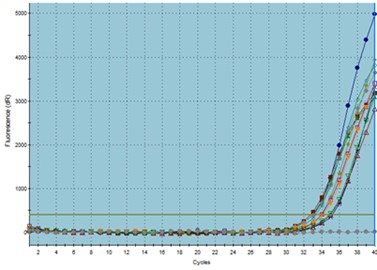
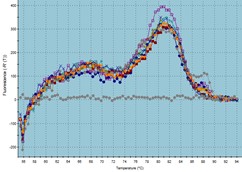
The amplification plot and melting curve of each sample were analyzed (Figure 1). Among the 25 patient samples, 20 samples were positive for the presence of C. difficile in the colonic bacterial population. In contrast, out of 25 samples from healthy individuals, five samples showed evidence of C. difficile. As a result, the ratio values indicated a higher frequency of C. difficile in the stool of CRC patients. Furthermore, the Chi-square test results revealed a significant difference between the patient and healthy groups in terms of the presence of C. difficile in the colon (P<0.001).
A
Figure 1. TaqMan real-time PCR assay: a) amplification plots, b) melting curves
The colorectal cancer is rapidly growing as a significant health concern in both developed and recently industrialized countries in a relatively brief time frame. This condition places a considerable burden on the healthcare systems worldwide, particularly affecting the well-being of young patients. The lifestyle plays a significant role in disturbing the normal balance of the gut microbiota (20, 21). Moreover, the interplay between the immune system and microbiota significantly influences the governance and control of mucosal immunity. The observed dysregulation in mucosal immune responses in the CRC patients may be associated with dysbiosis in the gut microbial communities (22, 23). Dysbiosis could be attributed to a decrease in the diversity of the normal flora of the gastrointestinal tract and the colonization of pathogenic or opportunistic bacteria (24, 25).
Recent studies have revealed that patients treated with broad-spectrum antibiotics, particularly those hospitalized and immunocompromised, demonstrate an elevated risk of developing CDI. Considering the existence of these risk factors in the CRC patients, there is a great susceptibility to the rapid development of CDI in the CRC patients (26). Persistent inflammation caused by CDI could be one of the pathways leading to the initiation of CRC through DNA damage (27).
In this study, real-time PCR was used to investigate the presence of C. difficile strains in the colons of CRC patients and healthy volunteers. The results indicated a significant difference between the CRC patients and healthy individuals in terms of the presence of C. difficile in the colon environment.
Numerous studies conducted inside and outside our target community using similar methodologies have also yielded similar results. For instance, Fukugaiti et al. (2015) employed real-time PCR in a study conducted in Brazil, revealing a substantial variation in C. difficile abundance between CRC and healthy groups (28). This finding aligns with the present study results, supporting the notion that C. difficile presence is distinguishable in individuals with CRC compared to those without CRC. In the Philippines, an ELISA-based investigation identified elevated levels of anti-C. difficile antibodies in individuals with CRC compared to the control group (9). This observation is also in line with the present study findings, supporting the idea that there is a significant correlation between C. difficile and CRC. This is evidenced by distinct immunological responses in the affected individuals. Furthermore, a study conducted in China analyzed stool cultures from the CRC patients and found notable differences in the intestinal carriage of C. difficile between the CRC and control groups. There was no apparent association with hospitalization (26).
Although distinct in methodology, the present study findings resonate with these results, collectively emphasizing the importance of C. difficile presence in the context of CRC. Furthermore, a cohort study conducted on a large population, which differentiated between C. difficile positive and negative groups, found that individuals in the former group were approximately 2.7 times more likely to be diagnosed with CRC (29). This observation reinforces the present study results, highlighting the epidemiological importance of C. difficile in the development of CRC. The impact of C. difficile on CRC is not yet fully understood. However, the hospitalized patients with CRC often exhibit numerous risk factors associated with C. difficile. Drewes et al., (2022) showed that C. difficile isolated from a CRC patient had tumorigenic potential in mice. The study elucidated that this bacterium induced tumorigenesis by triggering the Wnt signaling pathway and elevating the generation of IL-17-producing cells. This insight offers a mechanistic understanding of the possible connection between C. difficile and CRC, reinforcing the importance of the present study findings, highlighting the clinical implications of C. difficile presence (30).
These findings provide a strong basis for understanding the complex relationship between C. difficile and CRC, highlighting the potential diagnostic and therapeutic implications of C. difficile in this clinical context.
In addition to the significant findings indicating the presence of C. difficile in the CRC patients compared to the healthy controls, it is crucial to acknowledge the potential influence of confounding variables on our results. In particular, the impact of medication usage, particularly broad-spectrum antibiotics, cannot be overlooked, as they have been shown to disrupt gut microbiota composition and promote dysbiosis. Despite the exclusion criteria for the study subjects indicating that they should not have reported the use of antibiotics in the last three months, there is still a paucity of information regarding their previous infectious diseases that may have led to the long-term antibiotic use. As a consequence of this limitation, the definitive interpretation of the observed difference in the presence of C. difficile between the CRC group and the healthy controls is constrained. Moreover, unmeasured variables, such as genetic risk factors, may also contribute to the association between C. difficile and CRC progression, and further underscoring of the complexity of gut microbiota interactions in CRC. Future studies should aim to address these limitations by collecting comprehensive clinical and demographic data in order to better elucidate the role of C. difficile in the development and progression of colorectal cancer.
Similar to other studies, the present research indicated a higher incidence of C. difficile in the colon contents of the CRC patients compared to the healthy individuals.
Therefore, early identification and management of CDI are crucial and challenging aspects of the CRC patient care. However, additional research is necessary to determine the precise risk factors that contribute to the progression of C. difficile colonization to the active CDI in individuals with CRC.
We are grateful to the Research Council of the Tarbiat Modares University and the Tropical Infectious Diseases Research Center for their support on the project.
Ethical Considerations
All participants in this study signed an informed consent form. All protocols related to this research were approved by the Research Ethics Committee of Tarbiat Modares University (ethics code: IR.MODARES.REC.1399.083).
Authors’ Contributions
B.B, M.S.F and F.F: study idea, study design, supervising the work. Writing the manuscript. M.S.F & T.Sh: sample collection. B.B, F.F & T.Sh: molecular techniques and writing/editing the manuscript. A.R: data analysis. All authors read and approved the final version of the manuscript.
None.
Conflicts of Interest
The authors declare no conflict of interest.
Received: 2024/03/3 | Accepted: 2024/05/16 | ePublished: 2024/05/25
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |