BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-2323-en.html
The virulence factors are the pathogenicity degrees of the bacteria to the organisms, which determine the capability of the bacteria to introduce itself within the host. They give specific characteristics to the pathogen, facilitate the way to threaten the natural defense of the host, and thus cause a disease (1, 2). The most important virulence factors are antibiotic resistance, bacterial toxins, and various enzymes, which help the pathogen to familiarize with different environments. The effects of pathogenic bacteria in their hosts rely on the bacterial virulence factors, which assist them to survive and become virulent. Bacterial virulence factors range from the membrane to the secretory proteins, toxins, and exoenzymes such as lipase, protease, and hemolysin (3). Microbial virulence factors include a wide variety of the molecules produced by the pathogenic microorganisms, boosting their ability to escape their host defense system and cause disease (4).
The bacterium S. aureus secretes a number of enzymes such as lipase, protease, and hemolysin that help to spread bacteria in the body. The pathogenicity of S. aureus is caused by the secreted virulence factors such as proteases and host protease modulators. These virulence factors stimulate adhesion and invasion of bacteria through damage of tight junction barriers and keratinocytes.
The K. Pneumonia and P. mirabilis are both Gram-negative, rod-shaped bacteria that belong to the Enterobacteriaceae family. They are common cause of urinary tract infections, particularly in individuals with underlying urological abnormalities. They are opportunistic pathogens and cause several diseases in the human body. They make capsule, which consists of a polysaccharide substance and plays a role in hindering the process of phagocytosis by hiding the bacteria that may cause bacterial flakes in the urinary tract system. They can cause severe pneumonia, especially in immunocompromised individuals or those with underlying conditions like chronic obstructive pulmonary disease (COPD). It secrets persistent and heat sensitive disease (5).
The S. pyogenes bacteria are Gram-positive, chain-forming bacteria that commonly colonize the upper respiratory tract and skin. They are animal models for certain pathologies. The bacterium has evolved into a substantial virulence factor, enabling it to survive and proliferate in the human host. Some of these factors are important for adhesion, nutrient uptake, modulation of coagulation, etc. The virulence factors can alter the immune system detection of and response to the infections (6). The present study aimed to determine some enzymes (lipase, protease, hemolysin) as virulence factors for some Gram-positive and Gram-negative bacteria from different source samples.
The samples (80) were collected from various clinical sources of all genders and ages ranging from 1-80 years from Al-Haboubi Hospital in the city of Nasiriyah, Iraq from June 2022 to December 2022. The samples were transferred to the laboratory in the Department of Life Sciences/College Education of Pure Science using the sterile cotton swabs and placed in the incubator at 37°C for 2-4 hr. The clinical samples included nasal, ear, eye, and urinary tract infection (UTI) swabs. The samples were characterized phenotypically for their presumptive identification. The grouping was done based the cell morphology, colony morphology, Gram reaction, as well as Vitek 2 Compact system (7). The physiological tests including growth at different pH, temperatures, and salt tolerance were performed (8). The biochemical characterization of the isolates such as sugar fermentation test, nitrate reduction test, urease test, and IMViC (Indole, Methyl red; Voges-Proskauer and Citrate) test were also performed specifically for the Gram-negative isolates (9).
Detection of hemolysin was done according to the detection of cell-secreted virulence factor; haemolysin production using blood agar media (10). The plates were incubated at 37°C for 24 hr and then checked for the haemolysis. The presence of a clear zone around the colonies was taken as positive for the haemolysin production.
The extracellular protease production was detected according to the protease activity. The skimmed milk agar was used to assess the isolates for their ability to produce protease enzymes (casein hydrolysis) as described before (11). The isolates were streaked on the 2% skimmed agar and incubated at 37ºC for 24 hr, plates were checked for halo-regions around the streaks. A zone of clearing more than 1 mm around the streaks was recorded as positive for the protease production.
Lipase production was detected according to previous report (12). Briefly, the egg yolk agar plates were inoculated by the isolates and incubated at 37ºC.
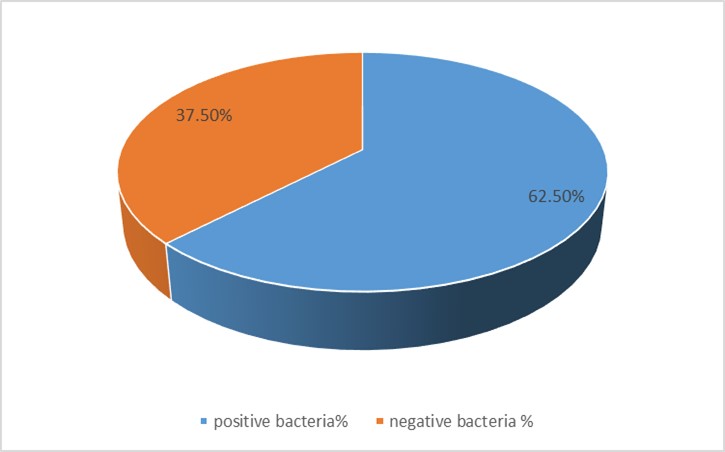
The samples were collected from various sources (ear, throat, eye, UTI, pharynx) and identified based on their microscopic, cultural, and biochemical characteristics as shown in Table 1. The percentage of Gram-positive and Gram-negative isolates from different sources are shown in Figure 1.
The results showed higher percentage (62.50%) of Gram-positive bacteria compared to the Gram-negative bacteria (37.50%) (Figure 1). The Gram-negative bacteria contain an important factor in their virulence that is the wall that contains a thin layer of peptidoglycan and a layer of lipoproteins and many liposaccharides and phospholipids. The identified Gram-negative bacteria included S. pyogenes, S. aureus, and klebsiella proteus that produce a variety of enzymes, which cause damage to the host tissues (13).
Table 1. The number and percentage of different sources of isolates.
| Present % | numbers | Source of isolate |
| 25.00% | 20 | Ear |
| 18.75% | 15 | Throat |
| 22.50% | 18 | Eye |
| 13.75% | 11 | Pharynx |
| 20.00% | 16 | UTI |
| 100% | 80 | Total |
Figure 1. Percentages of the isolated Gram-positive and Gram-negative bacteria from different sources
The results showed higher percentage (62.50%) of Gram-positive bacteria compared to the Gram-negative bacteria (37.50%) (Figure 1). The Gram-negative bacteria contain an important factor in their virulence that is the wall that contains a thin layer of peptidoglycan and a layer of lipoproteins and many liposaccharides and phospholipids. The identified Gram-negative bacteria included S. pyogenes, S. aureus, and klebsiella proteus that produce a variety of enzymes, which cause damage to the host tissues (13).
Bacterial secretion systems are important factors to determine the pathogenicity of the bacteria. They are crucial for the bacteria interaction with the host cells. The Gram-positive bacteria have a cytoplasmic membrane and a very thick layer of peptidoglycan. However, Gram-negative bacteria have the cell wall around the outer and cytoplasmic membranes that is a thin layer of peptidoglycan. Some of the Gram-negative and Gram-positive bacteria contain capsules. As the cell wall structures are complex and rigid necessary mechanisms are made for the proteins translocation across the membranes. These secretion systems include secretion of toxins or enzymes into the extracellular environment, and the transport of proteins that make pili or flagella.
The results of the study showed 31 (38.75%) isolates belong to S. aureus, while other Gram-positive isolates showed 19 (23.75%) for S. pyogenes. Gram-negative bacteria were 17 (21.25%) isolates for K. pneumonia and 13 (16.25%) isolates for P. mirabilis.
The ability of the isolates to produce hemolysin was tested by culture in a blood culture medium containing 5% human blood. The results showed that the Staphylococcus aeruse, K. Pneumoniae, Streptococcus, and P. mirabilis isolates produced the enzyme hemolysin. The results of some pathogencity factors of the isolates like their ability of produce homolysin indicate that all isolates were hemolysin producers (100%). Our results were similar to what was found before (13). The enzyme hemolysin, which is one of the main virulence factors, is one of the important factors produced by bacteria to obtain the iron. Hemolysin is an exotoxin that destroys the cell membrane of red blood cells. It is secreted by Gram-positive and Gram-negative bacteria both (14).
Microbial proteases that breakdown proteins are considered as virulence factors to cause several diseases. Interestingly, these enzymes are known to cause damage to the host organism defensive proteins. Proteases are known as antimicrobial peptides (15). They are partly determined by exo-products like and elastase and alkaline protease. They are responsible for the tissue damage by degrading elastin, collagen, and proteoglycans. These enzymes also degrade proteins that function in the host defense in vivo (16). The results were in agreement with the previous researchers’ results (17), who found similar and higher results of proteases. All isolates of this study were P. mirabilis, K. pneumonia, S. pyogenes, and S. aureus
Lipase is an enzyme that catalyzes the hydrolysis of oils and fats into the free fatty acids, monoglycerides, diglycerides, and glycerol. The results were in agreement with previous studies results (18). The enzyme lipase digests the host cellular lipid for its nutritional demand. Phospholipase is an enzyme released by bacteria with the ability to destroy host tissues (19). Study of lipase showed lipase production from Klebsiella, S. pyogenes, and P. mirabilis, however, S. aureus isolates according to the results, were proteolytic rather than lipolytic.
The presence of various virulence factors increases the pathogenicity ability of bacteria to evade defense approaches activated by the host leading to the therapeutic challenges. Among different virulence factors detected in K. pneumoniae, S. aureus, S. pyogenes, and P. mirabilis isolates from different sources, protease and hemolysin, were found to be the predominant virulence factors.
We acknowledge the clinical staff, who facilitated the sample collection. Special appreciation goes to the Department of Biology and Education Collage of Pure Science for their invaluable technical support.
Ethical Considerations
The study was conducted in compliance with the principles established in the "Helsinki Declaration” and authorized by the Education Collage of Pure Science, University of Thi-Qar's in-house Ethics Committee (Ref-50, Date: 18/03/2024).
Authors’ Contributions
Sanaa Mahdi Oraibi: conceptualization, data curation, formal analysis, investigation, methodology, resources, writing original draft, writing review & editing.
This study was self-funded.
Conflicts of Interest
Received: 2024/05/1 | Accepted: 2024/08/6 | ePublished: 2024/08/18
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |