BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-2267-en.html


 , Houda Benaicha2
, Houda Benaicha2 
 , Laila Reklaoui1
, Laila Reklaoui1 

 , Rajae Alloudane1
, Rajae Alloudane1 
 , Said Barrijal3
, Said Barrijal3 


2- Institut Supérieur des Professions Infirmières et Techniques de Santé de Tanger, Tangier, Morocco
3- Laboratory of Biotechnological Valorization of Microorganisms, Genomics, and Bioinformatics, Faculty of Sciences and Techniques, Abdelmalek Essaadi University, Tangier, Morocco ,
The emergence of multidrug-resistant (MDR) bacteria presents a significant concern for global public health. This emergence is constantly increasing, further exacerbating the healthcare system and potentially resulting in a therapeutic impasse (1). Gram-negative bacteria (GNB) are the most acknowledged organisms for their ability to resist multiple antibiotics. The antibiotic resistance in GNB is mainly due to their distinctive structure, particularly their outer cell membrane, which serves as an extra shield preventing antibiotics from reaching their intended targets. Noteworthy members within this group include the Enterobacteriaceae family, which includes Escherichia coli (E. coli), Klebsiella pneumoniae (K. pneumoniae), Enterobacter spp, Citrobacter freundii (C. freundii), and Proteus mirabilis (P. mirabilis) (1, 2). Infections caused by this particular group of bacteria are generally treated by antibiotics such as carbapenems and cephalosporins (3). However, the emergence of MDR bacteria worldwide and the lack of novel therapeutic alternatives have compelled healthcare professionals to reintroduce some old molecules, often discarded owing to their toxicity (3, 4). One such molecule is colistin (polymyxin E), an antibiotic that was discarded from clinical practice in the 1980s due to its neurotoxicity and nephrotoxicity, in preference for novel antibiotics such as third-generation cephalosporins and large-spectrum beta-lactams (3-5). Consequently, since 2012, colistin has been prescribed as an antibiotic to treat severe multi-drug resistant GNB-associated infections. Colistin is mainly administered intravenously (IV), although it has been shown that inhaling colistin can also be an effective treatment for respiratory infections. Interestingly, colistin inhalation proved lower toxicity compared to IV administration (4-6). However, the excessive consumption of colistin in recent years has resulted in the emergence of bacteria that resist this antibiotic. The resistance to colistin can be categorized into two types: chromosomal resistance and plasmid-mediated resistance. Chromosomal resistance originates from genetic mutations in the genes encoding the two-component systems (TCS) PhoP/phoQ and PmrA/PmrB, among others, subsequent to prolonged use of colistin (7). On the other hand, plasmid-mediated resistance results from acquiring plasmids that harbor mobile colistin resistance genes (mcr) (7). A significant number of studies have investigated into the dissemination of these plasmids in various samples worldwide, including humans, animals, food, and the environment (7). Consequently, ten mcr-like genes (mcr-1 to mcr-10) have been reported. These genes have been detected in most enterobacteriaceae strains, as well as Pseudomonas spp. and Acinetobacter spp (8, 9).
The detection of the mcr-1 gene in clinical isolates for the first time in Morocco is a worrying issue due to the high transferability of these plasmid genes (8, 9).
This study aimed to investigate the prevalence of colistin resistance and mcr genes among clinical Enterobacteriaceae isolated from the northwest of Morocco.
Bacterial Isolates
In total, 338 clinical isolates (from non-hospitalized patients) of Enterobacteriaceae were collected between December 2020 and May 2021 in North-West Morocco from medical analysis laboratories. The isolates were obtained from urine specimens and identified using a VITEK®2 system (Biomerieux, France). The isolates included 263 E. coli, 54 K. pneumoniae, 10 Enterobacter spp, 7 P. mirabilis, and 4 C. freundii. We had access to information regarding the patient's gender for 164 isolates out of 338 collected. The available data included 108 males, 44 females, and 12 children.
Antimicrobial Susceptibility Testing (AST)
Susceptibility testing was evaluated for all isolates using disc diffusion and minimum inhibitory concentration (MIC) following Clinical and Laboratory Standards Institut (CLSI) recommendations (10, 11). Nine antibiotics were tested for disc diffusion: Amoxicillin (AMX=30 µg), Cefotaxime (CTX=30 µg), Cephalotin (CF=30 µg), Ceftazidime (CAZ= 30 µg), Imipenem (IMP=10 µg), Nalidixic Acid (NA30 µg), Ciprofloxacin (CIP=5 µg), Tobramycin (TOB=10 µg), and Amikacin (AMK=30 µg). E. coli ATCC® 25922 was used as a quality control reference strain.
Unlike other antibiotics, the disc diffusion method is not recommended for colistin susceptibility determination (11). Alternatively, minimum inhibitory concentration (MIC) was determined using the broth microdilution method following CLSI recommendations (10). Briefly, 50 μl of each isolate was adjusted at optical density (OD) at λ = 600 nm (OD600) = 0.01 (5.105 colony-forming units (CFU)/mL) in cation-adjusted Mueller Hinton broth (Sigma–Aldrich, France) and added to 50 μl of colistin (Colistin sulfate salt, Sigma-Aldrich, France) (diluted in distilled water) at concentrations ranging from 0.5 to 64 µg/mL in 96-well plate. The mixture was incubated for 24 h at 37°C with shaking at 120 rpm. Subsequently, 10 µL of resazurin (Sigma-Aldrich, France) (0.015% (v/v)) was added to each plate's well to determine the MIC. The color change from purple to pink indicated bacterial growth. The MIC was determined as the lowest concentration at which no color change was observed (12). The isolates were considered susceptible to colistin when their MIC ≤2 µg/mL (10). Each strain that resisted at least three classes of antibiotics was considered an MDR strain (13).
Plasmid DNA Extraction and Quality Control
All isolates that were found colistin-resistant in the susceptibility testing were subjected to plasmid DNA extraction using a plasmid kit (GeneJET Plasmid Miniprep Kit, Thermo Scientific™, USA). Subsequently, the extracted DNA's purity (260/280 ratio) and concentration were measured using a nanodrop spectrophotometer (NanoPhotometer® P300-Class, Germany). The purified DNA was aliquoted and stored at –20°C.
PCR Amplification of Mcr-1, 2, 3, 4, 5 Genes
Genes mcr-1 to mcr-5 were amplified using the pairs of primers listed in Table 1. The PCR reaction mixture was prepared in a final volume of 50 μl including 10 µL of 5x MyTaq reaction buffer, 5 µL of DNA template, 1 µL of 20 mM of each primer, 1 µL of MyTaq DNA polymerase (Bioline, UK), and water (ddH2O) up to 50 µL. Thermal cycles were conducted using a thermal cycler (MultiGene™ Mini Personal, USA) (14-18). The amplicons were separated on electrophoresis (Mini-Sub Cell GT Systems, USA) using 1% agarose gel (for 40 min) containing 0.5 μg/mL ethidium bromide (Sigma–Aldrich, France). The bands were visualized using a gel imaging system (Quantum-ST4-1120/Blue, Spain) and compared to a DNA ladder (HyperLadderTM 100bp, Bioline, UK) to determine the amplicon's size. Colistin susceptible strain E. coli ATCC® 25922 was used as negative control.
Table 1. Primers used for PCR amplification of mcr-1, mcr-2, mcr-3, mcr-4, mcr-5 genes.
| Genes | Primer Sequence (5ʹ→ 3ʹ) | Size (bp) | Annealing Temperature (°C) | References |
| mcr-1 F mcr-1 R |
CGGTCAGTCCGTTTGTTC CTTGGTCGGTCTGTAGGG |
309 | 51 | (14) |
| mcr-2 F mcr-2 R |
TGTTGCTTGTGCCGATTGGA AGATGGTATTGTTTGGTTGCTG |
567 | 58 | (15) |
| mcr-3 F mcr-3 R |
TTGGCACTGTATTTTGCATTT TTAACGAAATTGGCTGGGTGGAACA |
542 | 50 | (16) |
| mcr-4 F mcr-4 R |
ATTGGGATAGTCGCCTTTTT TTACAGCCAGAATCATTATCA |
487 | 54 | (17) |
| mcr-5 F mcr-5 R |
ATGCGGTTGTCTGCATTTATC TCATTGTGGTTGTCCTTTTCTG |
1644 | 54 | (18) |
The PCR products were subjected to the Sanger sequencing method at the National Center for Scientific and Technical Research (NCSTR) in order to confirm the presence of the mcr-1 gene. The sequencing results were analyzed using SnapGene viewer software (Version 6.0), and the sequences were compared to those in the National Center for Biotechnology Information (NCBI) GenBank®.
Antimicrobial Susceptibility Testing (AST)
For 338 clinical isolates, we noted that AMX exhibited the lowest efficacy, with a resistance rate of 68.93%. Conversely, the antibiotic with the highest effectiveness against these isolates was IMP, with a susceptibility rate of 96.75%. The resistance rates of the isolates are summarized in Table 2.
For colistin, the microdilution method revealed that 15.08% (51/338) of the isolates were resistant to colistin. The MIC values for the isolates resistant to colistin ranged from 4 µg/mL to ≥ 64 µg/mL (Table 2).
Additionally, the present study revealed that of the total 338 isolates involved in this study, 77 (22.78%) were multidrug-resistant (MDR).
Occurrence of Mcr-Like Genes Among the Colistin Resistant Isolates
The 51 isolates that were found resistant to colistin (MIC ≥ 4 µg/mL) were subjected to PCR technique for screening the presence of mcr-genes (from mcr-1 to mcr-5).
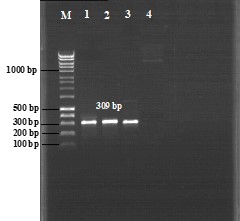
The results revealed that 3 of the 51 isolates (5.88%) harbored the mcr-1 gene (Figure 1), whereas no other isolate carried mcr-2 to mcr-5 genes.
All three isolates that were tested positive for mcr-1 gene were E. coli isolates. The description of the isolates carrying mcr-1 gene is summarized in Table 3.
Figure 1. Mcr-1 gene electrophoresis gel (309 bp). M, 100 bp DNA marker. Lane 1, E. coli TT-7. Lane 2, E. coli TT-48. Lane 3, E. coli TG-12. Lane 4, Negative control (E. coli ATCC® 25922).
Table 2. Resistance rates of the clinical isolates.
| E. coli n=263 (%) |
K. pneumoniae n=54 (%) |
P. mirabilis n=7 (%) | Enterobacter. spp n=10(%) | C. freundii n=4 (%) |
Total n=338(%) | |
| AMX | 165(62.73%) | 54(100%) | 7(100%) | 3(30%) | 4(100%) | 233(68.93%) |
| CF | 133(50.57%) | 32(59.25%) | 4(57.14%) | 3(30%) | 3(75%) | 175(51.77%) |
| CXT | 33(12.54%) | 6(11.11%) | 1(14.25%) | 2(20%) | 2(50%) | 44(13.01%) |
| CAZ | 39(14.82%) | 14(25.92%) | 1(14.25%) | 4(40%) | 0(00%) | 58(17.15%) |
| IMP | 6(2.28%) | 3(5.55) | 2(28.57%) | 0(00%) | 0(00%) | 11(3.25%) |
| TOB | 46(17.49%) | 8(14.81%) | 1(14.25%) | 3(30%) | 0(00%) | 58(17.15%) |
| AMK | 32(12.16%) | 5(9.25%) | 2(28.57%) | 0(00%) | 1(25%) | 40(11.83%) |
| NAL | 85(32.31%) | 18(33.33%) | 2(28.57%) | 4(40%) | 2(50%) | 111(32.84%) |
| CIP | 64(24.33%) | 15(27.77%) | 1(14.25%) | 3(30%) | 0(00%) | 83(24.55%) |
| COL | 33(9.76%) | 11(20.37%) | 7(100%) | 0(00%) | 0(00%) | 51(15.08%) |
AMX: Amoxicillin, CF: Cephalotin, CXT: Cefotaxime, CAZ: Ceftazidime, IMP: Imipenem, TOB: Tobramycin, AMK: Amikacin, NAL: Nalidixic Acid, CIP: Ciprofloxacin
Table 3. The description of the isolates carrying mcr-1 gene.
| Isolates reference | Strain | City | Origin of samples | Sex | Age (Year) | Colistin MIC (µg/mL) | Resistance profile |
| TT-7 | E. coli | Tetouan | Urine | male | 91 | 4 | AMX NA CPR TOB COL |
| TT-48 | E. coli | Tetouan | Urine | - | - | 32 | AMX, CLT, COL |
| TG-12 | E. coli | Tangier | Urine | male | - | >32 | COL |
Sequence Analysis
The PCR products were sequenced and found to be 99–100% identical to the mcr-1 genes available in the NCBI GenBank®.
The widespread availability and misuse of antibiotics, including self-medication and inadequate prescriptions, have caused microorganisms to develop self-defense mechanisms against antibiotics (19). The current situation poses a significant threat as it may deprive mankind of a critical weapon in their fight against pathogens. Consequently, We are on the verge of returning to a pre-antibiotic era, wherein even minor infections resulted in death (19, 20).
The most common pathogen in our study was E. coli, accounting for 77.81% (263/338) of all isolates. The second most prevalent pathogen was K. pneumoniae, accounting for 15.97% (54/338) of isolates. The other pathogens ranged between 1.18% and 2.95% (Table 2). Indeed, previous studies have also shown that E. coli is the most common cause of community and nosocomial bacterial infections. Furthermore, E. coli is the primary cause of urinary tract infections (UTI) among the Enterobacteriaceae family (21, 22). In the present study, females exhibited a higher prevalence of UTI (108/164) compared to males (44/164) and children (12/164). Indeed, numerous investigations have suggested that females are more prone to UTIs than males (21, 22). This finding aligns with our study results. The reason for this gender difference can be attributed to the physiological differences between males and females (21, 22).
Enterobacteriaceae isolates have high resistance rates to AMX while simultaneously being susceptible to carbapenems, which is another member of the beta-lactams family (23). This resistance is likely due to the widespread use of AMX, as it exhibits low toxicity. However, in contrast to other beta-lactams, carbapenems are particularly effective against Enterobacteriaceae because they resist the majority of β-lactamase, enzymes that break down beta-lactam antibiotics (23). These findings explain, in part, the high level of resistance of the isolates to AMX in our study (Table 2).
Following the discovery of plasmids harboring genes that encode colistin resistance (pHNSHP45, IncX4, IncHI2, and IncI2) in Enterobacteriaceae strains from both humans and animals (7, 8), a new controversy has erupted within the scientific community. This is a matter of great concern since colistin is regarded as a last-resort antibiotic for treating severe infections caused by MDR Gram-negative bacteria when other antibiotics proved ineffective. The first instance of colistin resistance (CoR) in Enterobacteriaceae isolates was in Athens (Greece) in 2004, and ever since, CoR has spread globally (24).
In our study, we found that colistin resistance was predominantly observed in P. mirabilis, followed by E. coli, and K. pneumoniae, to the detriment of other strains (Table 2). These findings were similar to Javed et al. (25). The high prevalence of colistin resistance in P. mirabilis isolates (7/7) can be attributed to their inherent (intrinsic) resistance (26). Conversely, for E. coli and K. pneumoniae isolates, the high prevalence of colistin resistance can be related to their high predominance in UTIs (27).
Colistin resistance has been reported in numerous other studies conducted globally, including in Europe (28, 29), Africa (30), Asia (31), North and South America (32, 33), and Australia (34). These findings reflect the concerning upward trend of this emerging resistance. The most alarming finding in our study was that 15.68% (8/51) of the CoR isolates, including 5 E. coli, were MDR and highly resistant to colistin (MIC values greater than 32 µg/mL) simultaneously. This suggests finding more CoR MDR strains as the study progresses since E. coli is the most common pathogen in UTI infections (21, 22), This could lead to an increase in the emergence of pan-drug resistance, leaving healthcare professionals unsure of the available therapeutic options to combat these superbugs when one of the "last men standing" (Colistin) is unable to stop them.
The plasmid-mediated mcr-1 colistin resistance gene can be traced back to China (8). After this discovery, a flurry of investigations has ensued, shedding light on the potential risks associated with the spread of the mcr-like genes (7-9, 14-18).
Throughout our study, we have successfully identified three E. coli clinical isolates that harbor the mcr-1 gene. To the best of our knowledge, this is the first instance of the mcr-1 gene detected in clinical isolates of Enterobacteriaceae (E. coli) in Morocco. Previously, the mcr-1 gene was only found in E. coli isolates from poultry (broiler chicken) and Pasteurellaceae isolates from ruminants (35, 36). Furthermore, our study is the first to investigate both the prevalence of colistin resistance and the presence of mcr-like genes in clinical isolates. Additionally, all three mcr-1 positive isolates in this study were found in two cities in Northern Morocco - Tetouan and Tangier. These cities are near each other (less than 63 kilometers apart) and are known as tourist destinations. Furthermore, they are located near Europe, specifically Spain (less than 20 kilometers), where the mcr-1 gene was first reported (37). This discovery raises concerns about the potential outbreak and widespread dissemination of these genes throughout the country since the mcr-like genes (mcr-1 to mcr-10) are typically carried on conjugative plasmids, which opens the possibility of their transfer from one bacterium to another (9, 14-18).
Interestingly, none of the 51 CoR isolates in this study harbored the genes mcr-2, mcr-3, mcr-4, and mcr-5. These findings are consistent with some previous investigations demonstrating that these genes are commonly less predominant when compared to the mcr-1 gene (38, 39). However, the resistance of the rest of the isolates (the isolates that did not harbor mcr-1 genes) to colistin can be explained by different mechanisms, for instance, they might still have chromosomal mutations in genes coding for one of the two-component systems (TCS), PmrA/PmrB and PhoP/phoQ, potentially leading to resistance against colistin. Interestingly, the activity of TCS PhoP/phoQ is regulated by a transmembrane protein, mgrb. When TCS PhoP/phoQ activity increases, mgrb protein inserts into the bacterial membrane, downregulating PhoP/phoQ activity. However, any mutation in the mgrb gene increases TCS PhoP/phoQ activity and leads to colistin resistance development (5, 7). Additionally, the inactivation of genes involved in LPS biosynthesis (lpxA, lpxC, lpxD) or overexpression of some efflux pumps (AcrAB-TolC, MexXY/OprM, KpnEF, RosAB, and Rnd-type), could contribute to the resistance against colistin in these isolates. Finally, it remains possible that these isolates might harbor mcr-like genes other than mcr-1 to mcr-5 (mcr-6 to mcr-10), resulting in their resistance to colistin (5, 7).
Consequently, all forms of resistance we discussed above (except for efflux pumps mutations), including plasmid-mediated resistance (mcr-1 to mcr-10) and chromosomal mutations (TCS PmrA/PmrB and PhoP/phoQ, mgrb gene, among others), lead to the same outcome, which is the modification of lipid A (colistin target). This modification occurs through the addition of phosphoethanolamine (pEtN) and 4-Amino-4-deoxy-L-arabinose (L-Ara4N) via phosphoethanolamine transferase enzyme, thus resulting in resistance to colistin (5, 7).
The majority of studies tend to ignore the presence of mcr-like genes in P. mirabilis as it is naturally resistant to colistin. However, a few studies reported the presence of mcr-1 and mcr-2 in P. mirabilis samples from human and animal isolates (25, 40). Neglecting to screen for plasmid genes in P. mirabilis could result in a silent spread of mcr genes among these bacteria and subsequently to other bacterial species.
The first-time detection of the mcr-1 gene in clinical isolates of Enterobacteriaceae in Morocco may worsen the healthcare situation by promoting the rapid spread of this gene within bacterial strains across the country. Consequently, it is urgently required to implement effective infection control measures to stop the spread of mcr-like genes.
Not applicable.
Ethical Considerations
This study was approved by the Ethics Committee for Research of the Faculty of Sciences and Techniques, Abdelmalek Essaadi University, Tangier, Morocco (CERB) (IORG0008724) and adhered to the Declaration of Helsinki.
Conflicts of Interest
The authors declare no conflict of interest.
Authors’ Contribution
Z.E. and H.B. designed the study. Z.E. and L.R. collected the data an performed experiments. Z.E. and R.A. analyzed the data. Z.E. wrote the main manuscript. S.B., H.B. and R.A. reviewed the manuscript. Authors accepted the final version of this manuscript.
No funding was received for conducting this study.
Received: 2023/12/13 | Accepted: 2024/03/14 | ePublished: 2024/03/18
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |