BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-2179-en.html

 , Mardiastuti H. Wahid2
, Mardiastuti H. Wahid2 

 , Fadilah Fadilah3
, Fadilah Fadilah3 

 , Rike Syahniar4
, Rike Syahniar4 

 , Ening Krisnuhoni5
, Ening Krisnuhoni5 

 , Andi Yasmon6
, Andi Yasmon6 


2- Department of Microbiology, Faculty of Medicine, Universitas Indonesia-Cipto Mangunkusumo Hospital, Jakarta, Indonesia
3- Department of Medical Chemistry, Faculty of Medicine, Universitas Indonesia, Jakarta, Indonesia
4- Department of Microbiology and Parasitology, Faculty of Medicine and Health, Universitas Muhammadiyah, Jakarta, Indonesia
5- Department of Anatomic Pathology, Faculty of Medicine, Universitas Indonesia-Cipto Mangunkusumo Hospital, Jakarta, Indonesia
6- Department of Microbiology, Faculty of Medicine, Universitas Indonesia-Cipto Mangunkusumo Hospital, Jakarta, Indonesia ,
Helicobacter pylori, a gram-negative bacterium, can be found in several forms: spiral, coccoid, or bent rods (1). As a micro-aerophilic bacterium, H. pylori grows only in low oxygen concentrations and survives in a strong acid environment by producing a urease enzyme to catalyze the hydrolysis of urea contained in stomach fluid to ammonia (2). This bacterium was found in the human stomach in 1982 and is the cause of several gastric disorders (1, 3, 4).
Several countries in Southeast Asia have reported a high percentage of H. pylori infections, such as Myanmar (69%), Thailand (54%), and Malaysia (20%) (5). In Indonesia, based on an Indonesian Gastroenterology Association research report, the percentage of H. pylori infections was about 22.1% (6). One of the problems in the management of patients with H. pylori infection is the emergence of H. pylori resistance to several antibiotics, particularly the first-line drugs: amoxicillin, tetracycline, levofloxacin, clarithromycin, and metronidazole (7). Studies conducted in Indonesia also showed that metronidazole (46.7%) had the highest prevalence in antibiotic-resistant H. pylori infections, followed by clarithromycin (42.9%), levofloxacin (31.2%), amoxicillin (5.2%), and tetracycline (2.6%) (7). Several mechanisms underlie the resistance of H. pylori to metronidazole, including decreased drug activation due to mutations in the rdxA gene (7, 8).
The rdxA gene encodes the nicotinamide adenine dinucleotide phosphate (NADPH) protein, which is a cofactor in the reduction of 5-nitroimidazole, leading to the production of cytotoxic free-radical nitro compounds (9, 10). Mutations in the rdxA gene that change amino acids of the NADPH protein will impair the ability of NADPH to reduce 5-nitroimidazole, leading to resistance (11). Kwon et al. reported substitutions, insertions, and deletions of amino acids that have played a role in the process of metronidazole resistance in North American isolates (12). Some mutations causing amino acid substitutions at positions 16, 56, and 68 were associated with metronidazole-resistant H. pylori (13). Many researchers have reported that mutations in the rdxA gene causing amino acid changes in the NADPH protein were involved in metronidazole-resistant H. pylori (10, 13-15).
Culture and molecular methods (PCR and DNA sequencing) can be used to determine bacterial resistance. Culture methods are less sensitive because H. pylori is highly fastidious and can easily change into coccoid forms (1, 16). Alternatively, molecular methods can identify mutations in the H. pylori rdxA gene. To our knowledge, most mutation analyses of the rdxA gene were conducted on bacterial isolates grown on culture media. In this study, we amplified a fragment of the rdxA gene by PCR from gastric paraffin biopsies. This approach would be beneficial for laboratories that find it difficult to grow H. pylori on culture media because of its highly fastidious characteristics.
2-1- Design and Specimens
This study was an explorative study using 34 gastric paraffin biopsies. The paraffin biopsies were obtained from the archives of Cipto Mangunkusumo Hospital Jakarta, with the approval of the Ethics Committee of the Faculty of Medicine, Universitas Indonesia-Cipto Mangunkusumo Hospital with reference number 0124/UN2.FI/ETHI CS/2018.
2-2- DNA Extraction
DNA extraction from gastric paraffin biopsies was performed as reported in Menoni et al. (17) with modifications. Two paraffin blocks, 5-10 mm large, were placed in a 1.5-mL tube; 1 mL xylene was added and mixed, and the tube was incubated for 5 min at room temperature. The mixture was centrifuged at 13800 g for 2 min, and the supernatant was discarded. The pellet was washed by using 70% ethanol, and air-dried at room temperature for 10 min. The pellet was extracted by using a QIAmp DNA Mini Kit (Qiagen, Germany) in accordance with the manufacturer’s instructions, with a final volume elution of 50 µL. The purity of DNA extraction was determined by using NanoDrop 2000 (Thermo Scientific).
2-3- Detection of H. pylori by Real-time PCR
All H. pylori-positive gastric paraffin biopsies in this study were confirmed by the real-time PCR method as we reported previously (18). The primers (forward [5’-CTC ATT GCG AAG GCG ACC T-3’] and reverse [5’-TCT AAT CCT GTT TGC TCC CCA-3’]) and probes (FAM-ATT ACT GAC GCT GAT TGC GCG AAA GC-TAMRA) specific for H. pylori 16S rRNA were used as reported by Ramírez-Lázaro, Lario (19).
2-4- Amplification of the rdxA Gene DNA Fragment
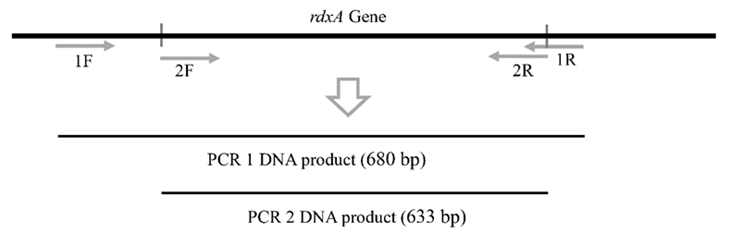
To increase the sensitivity, we conducted nested PCR (first PCR and second PCR) using two pairs of primers. PCR1 used 1F (5'-GTA TGC TAC GAA AAA TTC TA-3') and 1R (5'-TTG TTT AAT CAC AAC CAA G-3'), while PCR 2 used 2F (5'-ATG AAA TTT TTG GAT CA AG-3') and 2R (5'-TCA CAA CCA AGT AAT CGT A-3'). The primers were designed by using Designer Primer 2.0 software. The 1F and 1R primers were designed at upstream and downstream sites of the rdxA gene respectively, while 2F and 2R were designed for the full length of the rdxA gene (from start codon [ATG] to codon stop [TGA]) (Figure 1). The specificity of primers was analyzed by primer Blast in GenBank showing 100 % specificity for H. pylori and no cross-reaction with other bacteria (data not shown). PCR 1 and PCR 2 have product sizes of 680 bp and 633 bp, respectively. The compositions of PCR 1 (20 µL of reaction volume) and PCR 2 (40 µL of reaction volume) were 1x PCR Master mix (iNtRON BIOTECHNOLOGY, cat. no. 25266), 0.1 µM of each primer, and 4 µL (PCR 1) of the DNA extraction result or 2 µL (PCR 2) of the PCR 1 product. The thermal cycles (ABI GeneAmp™ system 2700) of PCR 1 and PCR 2 were performed under the following conditions: 94°C for 4 min; 40 cycles of 94°C for 30 sec, 52°C for 30 sec, and 72°C for 1 min; and 72°C for 5 min. The PCR 2 products were purified from gel agarose, and the pure DNA was directed to DNA sequencing by using 3500 Series Genetic Analyzers (Applied Biosystems, California, USA).
Figure 1. The PCR 1 using primers specific for upstream (1F) and downstream (1R) of the rdxA gene, respectively. PCR 2 using 2F and 2R primers for amplification of full length of rdxA gene. This illustration is not to scale.
The editing of overlapping DNA sequences was performed by SeqScape V2.7 (Applied Biosystems, California, USA). The analysis of 3D protein structure was conducted by using Robetta Software (https://robetta.bakerlab.org), and cavity binding activity was analyzed by Prankweb (https://prankweb.cz). Molecular docking was performed by MOE (Molecular Operating Environment) 2020 with the high-speed α-shape algorithm. MOE 2020 uses scoring terms such as London dG, FlexX, DrugScore, and Mcdock. A referral sequence of NADPH amino acid of metronidazole-sensitive H. pylori (wild type) obtained from GenBank with accession number: NC_000915.
Of the 34 H. pylori-positive paraffin biopsies (detection by real-time PCR), only 5 samples were amplified successfully for the rdxA gene by nested PCR (Figure 2). After the sequencing and editing process, we performed a GenBank Blast analysis of the DNA sequences, which reported the H. pylori NADPH protein with a range of percentage identity of 96.52-99.21%. The sequences have been deposited in GenBank with accession numbers OL874434, OM102363, OM102364, OM102365, and OM102366.
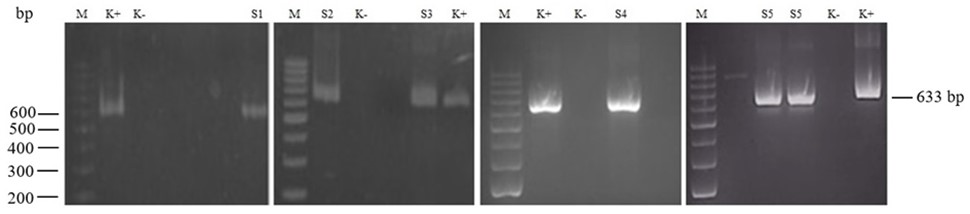
Figure 2. Nested PCR results for the H. pylori rdxA gene fragment with a 633-bp DNA length. Lane M: DNA ladder from 200 bp at the bottom to 1000 bp at the top. Lanes K+ and K-: positive and negative controls. Lanes S1-S5: PCR-positive ID samples. bp: base pair.
In all 5 gastric paraffin biopsies, H. pylori NADPH proteins showed amino acid substitutions at positions T31E, D59N, L62V, S88P, G98S, R131K, V172I, V204I, and A207T (Table 1). Two of the 5 samples showed nonfunctional proteins due to a premature stop codon (S4) and a frameshift (S5). Sample S4 demonstrated the presence of a premature stop codon, due to the substitution of C with T at position 31, thus forming a TAA stop codon. Sample S5 showed a frameshift due to the insertion of A at position 238, resulting in a shift in the reading of the codons and a stop codon at amino acid 97. On the other hand, sample S2 showed a different characteristic: the insertion of 6 DNA bases (TGGTAT) in the base positions 260-265, resulting in the addition of two amino acids, namely leucine (L) and valine (V), in base positions 87 and 88.
Table 1. Analysis of amino acid changes
| Strains | Positions of Amino Acid Changes | ||||||||||||||||||||
| 11 | 30 | 31 | 53 | 59 | 62 | 64 | 68 | 80 | – | – | 88 | 90 | 97 | 98 | 131 | 172 | 179 | 204 | 205 | 206 | |
| Wild type | Q | S | T | H | D | L | K | A | A | – | – | S | R | H | G | R | V | K | V | D | A |
| S1 | . | . | E | . | N | V | . | . | . | – | – | P | K | . | S | K | I | R | I | A | T |
| S2 | . | . | E | R | N | V | . | . | . | Lb | Vb | P | . | . | S | K | I | . | I | . | T |
| S3 | . | . | E | . | N | V | . | T | . | – | – | P | . | T | . | K | I | R | I | A | T |
| S4 | a | ~ | ~ | ~ | ~ | ~ | ~ | ~ | ~ | ~ | ~ | ~ | ~ | ~ | ~ | ~ | ~ | ~ | ~ | ~ | ~ |
| S5 | . | . | . | . | N | . | N | . | Frameshift | c | ~ | ~ | ~ | ~ | ~ | ~ | ~ | ||||
Dot (.): the same amino acids as in the wild type. Strip (–): insertion positions. a: a nonsense mutation in a nucleotide of codon 11 causing a premature stop codon. b: amino acid insertions. Frameshift: amino acid changes cause a change in the rdxA gene frame from codons 80 to 96. c: a premature stop codon caused by a frameshift. S1-S5: strains in this study. Wild type: NADPH amino acid sequence of metronidazole-sensitive H. pylori obtained from GenBank (accession number: NC_000915). Letter codes for amino acids: Q (Glutamine), S (Serine), T (Threonine), H (Histidine), D (Aspartic acid), L (Leucine), K (Lysine), A (Alanine), R (Arginine), G (Glycine), V (Valine), E (Glutamic acid), N (Asparagine), P (Proline), I (Isoleucine).
To determine the effect of the insertions of amino acids (sample S2) on the structure of the NADPH protein, we performed structure and docking analysis by comparing amino acid substitutions without (S2-S) and with insertions of two amino acids (S2-I) (Figure 3). Compared with the wild type, both S2-S and S2-I showed changes in amino acids forming the pocket structures of binding sites. S2-S showed two pockets that were larger than those of the wild type, while S2-I showed the loss of one pocket.
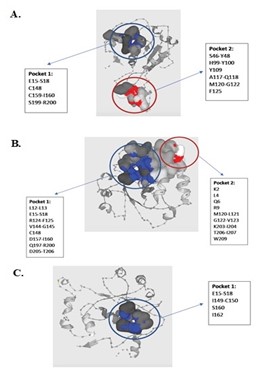
Figure 3. Analysis of 3D protein structure was conducted by using Robetta Software (https://robetta.bakerlab.org), and cavity binding activity was analyzed by Prankweb (https://prankweb.cz). A: NADPH protein structure of sensitive H. pylori (wild type) obtained from GenBank (accession number: NC_000915). B: NADPH protein structure of Strain S2 with substitutions (S2-S). C: NADPH protein structure of Strain S2 with both substitutions and insertions (S2-I). Text in boxes: amino acids with their positions. Pockets: active sites for the NADPH protein. Pocket 1 is indicated in blue, and pocket 2 in red.
Based on the docking analysis (Figure 4), the interaction of metronidazole with the wild type NADPH protein was affected by three amino acids (Ser18, Arg16, and Lys198) with -5.5243 kcal/mol. The interaction of metronidazole with the S2-S and S2-I NADPH proteins was affected by amino acids Ser130 and Arg126 (-3.2128 kcal/mol) and Arg10 (-2.8391 kcal/mol), respectively.
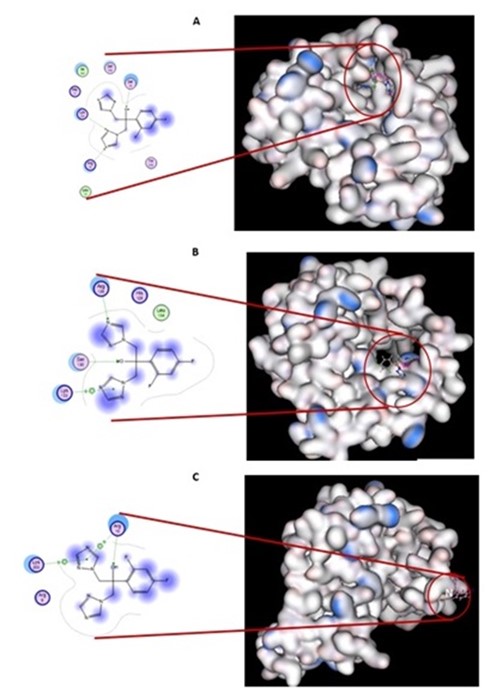
Figure 4. The 3D and 2D docking analysis between fluconazole and the NADPH protein. A: The NADPH protein of sensitive H. pylori (wild type) obtained from GenBank (accession number: NC_000915). B: the NADPH protein of Strain S2 with substitutions (S2-S). C: the NADPH protein of Strain S2 with both substitutions and insertions (S2-I). Amino acids in pink indicate a hydrogen bond acceptor, and amino acids in green a hydrogen bond donor.
For survival in an environment with the drug metronidazole, H. pylori has developed a major mechanism, namely mutations in the rdxA gene causing the impairment of the NADPH protein; these mutations mean that metronidazole is not active or cannot function optimally in damaging bacterial DNA (13). To determine the molecular characteristics of the NADPH protein, in this study we designed primers for nested PCR and DNA sequencing to amplify a fragment and to analyze the DNA sequence of the rdxA gene, respectively. Of the 34 H. pylori-positive paraffin biopsies, 5 could be used for the analysis of the rdxA gene. This indicates that the nested PCR had low sensitivity in amplifying the 633-bp rdxA gene fragment in paraffin biopsies. The amplification failure might be caused by degraded bacterial genomes in paraffin biopsies. Several studies have reported an effect of paraffin treatment on DNA degradation (20-22). Many factors can affect DNA degradation, including fixative compositions and types, treatment duration and temperature, and tissue type (20, 21, 23, 24).
The H. pylori NADPH proteins analyzed in this study showed several characteristics, namely substitutions, insertions, stop codons, and frameshifts with stop codons. Kwon et al. (12) reported that 7 out of 9 clinical resistant isolates had nucleotide additions (6 isolates) and mutations (1 isolate) causing frameshifts and premature stop codons (12). Mutations causing frameshifts and premature stop codons have also been reported by other researchers (25, 26). In this study, we found that 2 out of 5 strains had a premature stop codon caused by a nonsense mutation (Strain S4) and a frameshift and/or stop codon caused by a nucleotide insertion (Strain S5) (Table 1).
Regarding amino acid substitution, Mirzaei et al. (13) reported substitutions (R16H, R16C, S18F, H25R, M56I, A68T, A68V, H97Y, S108A, V111A, A118T, V204I, and W209R) that were detected only in resistant H. pylori isolates (13). In this study, we found the same substitutions (A68T, A68V, A118T, V204I) as reported by Mirzaei et al. (13) Of those substitutions, V204I was detected in 4 strains (S1-S4) (Table 1). Concerning the A118T substitution. Marques et al. (15) proposed that it is an important substitution due to the change from a non-polar amino acid (Ala [A]) to a polar non-charged amino acid (Thr [T]), and the substitution was detected with a higher frequency in resistant isolates (15). Moreover, Kim et al. reported that the amino acid substitutions in positions R10K, R16H, M21A, H53K, M56I, L62V, A68V, G98S, G163D, and A206T were detected in metronidazole-resistant strains (26). In this study, we found the same substitutions (L62V, A68V, G98S, A206T) as reported by Kim et al. Of those, 2 substitutions (L62V and A206T) were detected in 4 strains, and the others (G98S and A68V) were detected in 3 and 1 strain, respectively (Table 1).
Interestingly, we found 2 novel insertions in Strain S2 (Table 1). The structure of the protein was analyzed to determine the role of the insertions by comparing substitutions (S2-S) with both substitutions and insertions (S2-S). S2-S showed a change of amino acids forming the pocket structures of binding sites (Figure 3) and a decrease in the affinity for metronidazole (Figure 4). These changes might decrease the efficiency of the NADPH protein in reducing metronidazole, thus impairing metronidazole toxicity. We hypothesize that the loss of a pocket causes an extreme impairment of metronidazole toxicity.
The nested PCR and DNA sequencing using specific primers designed in this study can be used for the analysis of the H. pylori rdxA gene detected in gastric paraffin biopsies. We also found that substitutions can affect the amino acids forming pockets for NADPH active sites. Unexpectedly, we found 2 novel amino acid insertions causing the loss of one pocket of the NADPH protein and leading to the lowest affinity for metronidazole, thus extremely impairing the capacity of the NADPH protein to reduce metronidazole. However, the effects of these two novel insertions on antibacterial-resistant phenotype should be addressed in a future study.
We thank the Department of Anatomic Pathology, Faculty of Medicine, Universitas Indonesia-Cipto Mangunkusumo Hospital for providing gastric paraffin biopsies.
Ethical Considerations
This study was approved by the Ethics Committee of the Faculty of Medicine, Universitas Indonesia-Cipto Mangunkusumo Hospital, with reference number 0124/UN2.FI/ETHI CS/2018.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors’ Contribution
MW and AY contributed to the conception, design, methodology, formal analysis, and acquisition. They critically drafted the manuscript, revised the manuscript, gave their final approval, and agreed to be accountable for all aspects of the work ensuring integrity and accuracy. WF contributed to the formal analysis, experimental process, verification, preparation and writing of the original draft, and editing of the manuscript. F contributed to the design, software, formal analysis, methodology, and validation. RS contributed to the specimen collection process.
This study was funded by a PUPT grant with reference number 2670/UN2.R3.1/HKP05.00/2017 from the Ministry of Research, Technology, and Higher Education of the Republic of Indonesia.
Received: 2023/12/1 | Accepted: 2024/03/7 | ePublished: 2024/03/18
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |