BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-2139-en.html
2- Department of Biology, College of Science, University of Mosul, Mosul, Iraq
Intestinal parasitic infections are a global public health concern, particularly in developing countries (1). The World Health Organization (WHO) reports that approximately 3.5 billion people worldwide are infected with intestinal parasites responsible for significant morbidity and mortality (2). Intestinal parasitic infections pose significant health challenges worldwide, particularly in developing countries with inadequate sanitation and limited access to clean water (3, 4). Among the numerous parasitic pathogens affecting the gastrointestinal tract, E. histolytica and Giardia lamblia are two prominent causes of diarrhea and intestinal disorders (5, 6). These parasites are responsible for substantial morbidity and mortality, particularly in regions where socio-economic conditions hinder effective disease control and healthcare delivery (7, 8).
Iraq, a country with a history of conflict and limited resources, is confronted with a high burden of intestinal parasitic infections (9, 10). The prevalence of G. lamblia infections remains a pressing concern, especially among individuals presenting with diarrhea and intestinal disorders in hospitals across Iraq (11). Only a little data is available regarding the prevalence of E. histolytica. According to the study conducted in 2011 by Hussein et al. (12), the prevalence of E. histolytica among 730 fecal samples was 24.3%. It became over a decade; no studies have been performed on the prevalence and molecular detection of E. histolytica at the level of Iraq.
Conventional microscopy has long been the primary method for diagnosing intestinal parasitic infections (13). However, its limitations in terms of sensitivity and specificity have led to the development and adoption of molecular techniques for improved diagnostic accuracy (6). Polymerase chain reaction (PCR) and molecular-based assays offer enhanced sensitivity and specificity, enabling the detection of low-level infections and the differentiation of closely related species (14). Such molecular techniques have proven invaluable in parasitology, allowing for rapid and reliable detection of E. histolytica and G. lamblia (15).
Microscopy enables the direct visualization of parasites in clinical samples, allowing for immediate diagnosis (16), while PCR amplifies specific DNA sequences of the parasites, improving diagnostic sensitivity and providing species-specific information (17). Furthermore, gel electrophoresis facilitates the separation and visualization of PCR products, enabling the identification of subtypes within E. histolytica and G. lamblia populations (18).
The current study aims to investigate the prevalence and diagnosis of E. histolytica and G. lamblia infections among individuals suffering from diarrhea and intestinal disorders in hospitals throughout Iraq. The study employs a comprehensive approach utilizing conventional microscopy and molecular examination techniques, including PCR, to enhance diagnostic accuracy.
Study Design and Sample Collection
This study was a cross-sectional study conducted in hospitals in the city of Mosul, Iraq, between February 2022 and July 2023. Patients with diarrhea or other intestinal disorders who met the inclusion criteria (The inclusion criteria are people suffering from digestive disorders and diarrhea of varying severity, from the age of one year to the age of 70 years) were invited to participate in the study. Written informed consent was obtained from each participant or their parents/guardians if the participant was a child. A total of 371 stool samples were collected from consenting patients and transported to the laboratory in sterile containers.
Microscopic Examination
All stool samples were subjected to direct wet mount preparation, and saline and iodine mounts were used to detect the presence of E. histolytica and G. lamblia cysts and trophozoites. The samples were examined under a microscope by experienced technicians who were blinded to the participants' clinical information.
Molecular Examination
DNA was extracted from stool samples using a commercial kit (Qiagen, Germany) following the manufacturer's instructions. A set of primers was designed to amplify various fragments of gene specific for E. histolytica and Giardia lamblia (Table 1).
Table 1. The sequence of primers used for the detection of E. histolytica and Giardia lamblia
| Name of parasite | The primer | Sequence (5’-3’) | No. base pair | References |
| E. histolytica | UEH-F | TAAGATGCACGAGAGCGAAA | 20 | (19) |
| UEH-R | GTACAAAGGGCAGGGACGTA | 20 | ||
| Ehis-F | ATGCACGAGAGCGAAAGCAT | 20 | (20) | |
| Ehis-R | GATCTAGAAACAATGCTTCTCT | 20 | ||
| G. lamblia | AL3543 | AAATIATGCCTGCTCGTCG | 19 | (21) |
| AL3546 | CAAACCTTITCCGCAAACC | 19 | ||
| AL3544 | CCCTTCATCGGIGGTAACTT | 20 | ||
| AL3545 | GTGGCCACCACICCCGTGCC | 20 |
DNA amplification
PCR amplification was performed in a 20 µL reaction volume containing [10 µL 2X master mix, 1 µL Forward primer, 1 µL Reverse primer, 6 µL PRC grade water, 2 µL DNA sample]. The PCR conditions were as follows: initial denaturation at 95°C for 10 min, followed by 35 cycles of denaturation at 95°C for 1 min., annealing at [55°C for each of AL3543, AL3544, AL3545 and AL3546, while 58°C was used for UEH and 50°C for Ehis] for 45 s, and extension at 72°C for 1 min., with a final extension at 72°C for 10 min. The PCR products were visualized on a 1.5% agarose gel electrophoresis using a safe stain. The gel band for PCR product of E. histolytica samples was 166 bp while the band size for G. intestinalis was 530 bp.
Sequencing
A subset of PCR products (13 samples for each parasite PCR product) was selected for sequencing to confirm the accuracy of the PCR results. The PCR products were purified using a PCR purification kit (Qiagen, Germany), and sequencing was performed using an automated DNA sequencer (ABI 3730, Applied Biosystems, USA). The obtained sequences were compared with sequences available in public databases using BLAST software.
Phylogenetic analysis
The phylogenetic analysis assessed the genetic diversity and relatedness of E. histolytica and G. intestinalis isolates. The obtained DNA sequences, including the 16S rRNA gene for E. histolytica and the triose phosphate isomerase (TPI) gene for G. intestinalis, were aligned with reference sequences using nucleotide Blast in NCBI. Phylogenetic trees were constructed using the Maximum Likelihood method with the Tamura-Nei model in software such as MEGA11. Bootstrap analysis with 1000 replicates was performed to evaluate the robustness of the inferred phylogenetic relationships. The resulting trees were visualized using tree visualization software, and interpretation was based on the clustering and genetic distances between sequences.
Data Analysis
Data were analyzed using SPSS version 22 (IBM, USA). Descriptive statistics were used to calculate the prevalence of E. histolytica and G. lamblia infections. The chi-square test was used to compare the prevalence of infections between different groups. P-values less than 0.05 were considered statistically significant.
The results of laboratory tests for 371 collected samples showed that the percentage of infection with parasites was 212 samples, with a rate of (78%) for the amoeba parasite and (22%) for the giardia parasite. Among the infected samples, 166 samples were infected with E. histolytica, 46 samples were infected with G. lamblia, and 82 samples were co-infected with both parasites.
The prevalence of E. histolytica infection was significantly higher than that of G. lamblia infection (p < 0.001). The age and gender of the participants did not significantly affect the prevalence of infection with either parasite.
Sequencing results
Sequencing results for the TPI gene of G. intestinalis and 16S rRNA of E. histolytica isolates revealed the presence of the species in the samples. The obtained TPI gene sequences were submitted to the NCBI gene bank, and accession numbers were obtained for future reference (Figures 1 and 2).
To elucidate the genetic diversity and relationships of E. histolytica isolates from Iraq, sequencing of the 16S rRNA gene was performed. The resulting sequences were used to construct a phylogenetic tree using the Maximum Likelihood method based on the Tamura-Nei model in MEGA11 software. Bootstrap analysis with 1000 re-samplings was conducted to assess the robustness of the tree. Partial DNA sequences of concatenated partial 16S rRNA gene were utilized as input data for the phylogenetic analysis (Figure 3).
The phylogenetic tree of E. histolytica demonstrated the evolutionary relationships among the isolates from Iraq. The tree provided insights into the genetic diversity and relatedness of the identified E. histolytica isolates. The bootstrap values indicated the confidence level of the branching patterns within the tree.
(Figure 3) displays the phylogenetic tree of G. intestinalis based on the TPI gene sequences. The tree depicts the grouping of the identified G. intestinalis isolates from the study samples and their placement within the broader evolutionary context of G. intestinalis isolates worldwide.
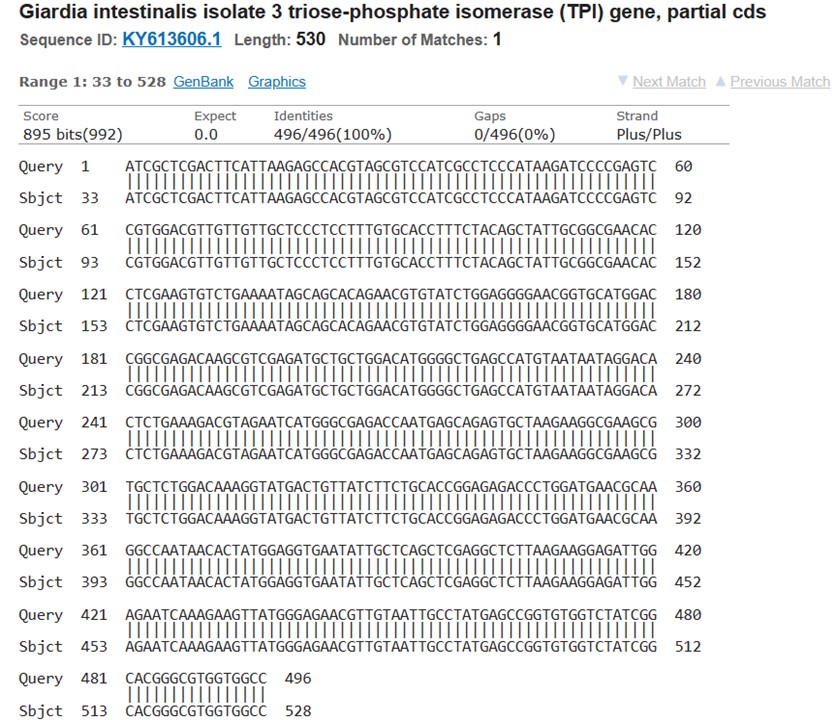
Figure 1. Sequencing results for the triose phosphate isomerase gene (TPI) of G. intestinalis
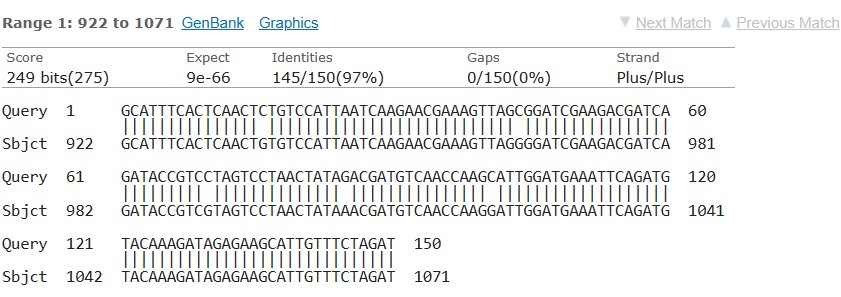
Figure 2. Sequencing results for the 16S rRNA for E. histolytica.
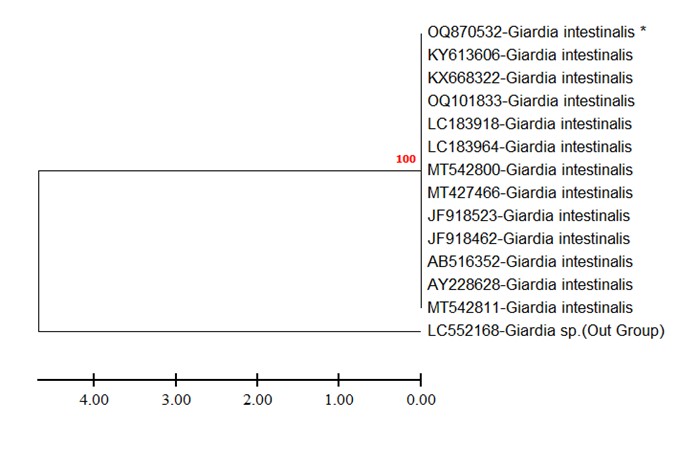
Figure 3. Phylogenetic tree of G. intestinalis based on TPI gene sequences
(Figure 4) shows the phylogenetic tree of E. histolytica from Iraq, based on the 16S rRNA gene sequences. The tree illustrates the clustering of the isolates from Iraq and their relationship with other known E. histolytica isolates from different geographic regions.
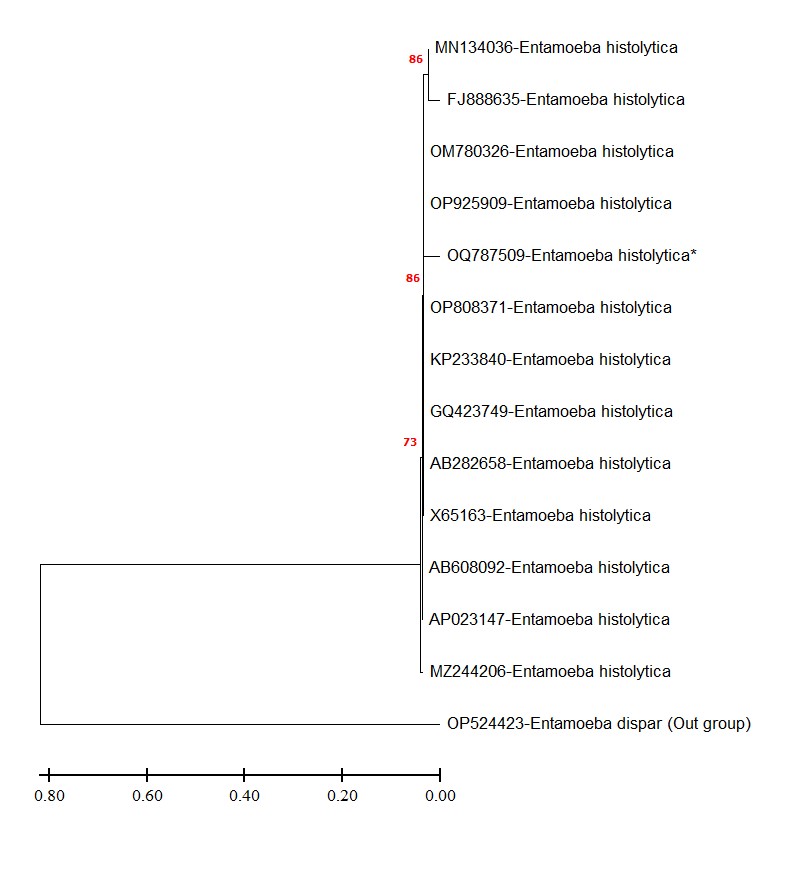
Figure 4. Phylogenetic tree of E. histolytica from Iraq based on 16S rRNA gene sequences.
The current study aimed to investigate the prevalence of E. histolytica and G. lamblia infections in patients with diarrhea and intestinal disorders in hospitals in Mosul, Iraq, using conventional microscopy and molecular techniques. The results showed that the prevalence of parasitic infections in the studied population was high, with E. histolytica being the most prevalent parasite. In this study, we employed molecular techniques, including sequencing and phylogenetic analysis, to gain insights into the prevalence, genetic diversity, and relatedness of these parasites among individuals presenting with diarrhea and intestinal disorders in hospitals across Iraq.
The prevalence of E. histolytica in this study was consistent with previous studies conducted in Iraq and other developing countries (22). About 48% of diarrheal patients were positive in Wasit governate-Iraq (23). Infection with Entamoeba species was recorded in Kurdistan region of Iraq in 21.68% (562/2592) of the analyzed studied specimens (24). Poor sanitation and hygiene practices are the main contributing factors to the high prevalence of E. histolytica infection in these areas. However, this study's lower prevalence of G. lamblia infection could be attributed to differences in the transmission routes and environmental factors affecting cyst survival (25).
Using molecular techniques such as PCR and sequencing has significantly enhanced the sensitivity and specificity of diagnosing parasitic infections. Moreover, combining microscopy and molecular techniques is essential for accurate diagnosis, particularly in cases where the parasite load is low (26).
The sequencing of the TPI gene for G. intestinalis confirmed the presence of this species in the studied samples. Our findings align with previous studies that have reported the high prevalence of G. intestinalis infections in Iraq (27). Identifying G. intestinalis using molecular methods such as sequencing provides accurate species-level determination, enabling a better understanding of the epidemiology and potential transmission sources (28). The availability of the obtained TPI gene sequences in the NCBI gene bank enhances the accessibility of this valuable data for future research and comparison with other global isolates.
The phylogenetic analysis determined the genetic characteristics of the parasite in order to discover the source of contamination in human societies. Previous studies in Iraq and neighboring countries like Iran revealed that the source is mostly zoonotic (29). Another study suggested that giardiasis disease is one of the endemic diseases in Iraq and the phylogenetic analysis showed that assemblage A was the most prominent type in Iraq (30). In comparison, no data was available for the previous decade for the Phylogenetic analysis of Entamoeba histolytica in Iraq.
The findings of this study have important implications for managing and preventing parasitic infections in Iraq. Health education programs aimed at promoting hygiene practices, improving sanitation, and providing safe water sources could significantly reduce the burden of parasitic infections in the population. However, the study has some limitations, including the possibility of selection bias as the study was conducted in hospitals, which may not reflect the prevalence of infections in the general population. Furthermore, the study did not evaluate other potential risk factors for infection, such as socioeconomic status, hygiene practices, or water and food sources.
In conclusion, this study provides valuable information on the prevalence of E. histolytica and G. lamblia infections in patients with diarrhea and intestinal disorders in hospitals in Mosul, Iraq. The study concluded that more than a quarter of the studied patients were positive for E. histolytica. The sequencing results confirmed the presence of these parasites, and the phylogenetic analysis provided insights into their genetic characteristics. These findings contribute to understanding the epidemiology and genetic diversity of these parasites in Iraq.
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Mosul University. Written informed consent was obtained from all participants, and confidentiality was maintained throughout the study.
'Not applicable.’
Conflicts of Interest
The authors declare any conflict of interest.
All authors contributed to data collection, writing, and manuscript revision. All authors read and approved the final manuscript.
'Not applicable.’
Received: 2023/06/1 | Accepted: 2023/07/18 | ePublished: 2023/07/26
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |








