BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-2067-en.html


 , Abbas Farahani2
, Abbas Farahani2 

 , Habibollah Turki1
, Habibollah Turki1 

 , Saeed Shoja3
, Saeed Shoja3 

 , Amin Ghanbarnejad4
, Amin Ghanbarnejad4 

 , Hamed Kamali1
, Hamed Kamali1 

 , Jebreil Shamseddin5
, Jebreil Shamseddin5 


2- Molecular and Medicine Research Center, Khomein University of Medical Sciences, Khomein, Iran
3- Infectious and Tropical Diseases Research Center, Hormozgan Health Institute, Hormozgan University of Medical Sciences, Bandar Abbas, Iran
4- Social Determinants in Health Promotion Research Center, Hormozgan Health Institute, Hormozgan University of Medical Sciences, Bandar Abbas, Iran
5- Molecular Medicine Research Center, Hormozgan Health Institute, Hormozgan University of Medical Sciences, Bandar Abbas, Iran ,
Blastocystis spp. is an obligate anaerobic protozoan parasite without a cell wall and has one or more nuclei around the central vacuole, rough and smooth endoplasmic reticulum, Golgi apparatus, and mitochondria-like organelles. The lack of growth in fungal environments and growth at 37˚C and normal pH, as well as resistance to antifungal drugs such as amphotericin B and sensitivity to metronidazole, placed it among protozoa (1-3). This parasite is very polymorphic and can be seen in several forms in direct smears and the culture medium. Generally, the parasite has four identifiable and main forms: Vacuolar, granular, amoeboid, and Cystic form. The formation of these shapes depends on factors such as osmotic changes, environmental conditions, metabolic status, and internal and external physical factors (4). After long controversial evidence, organisms were classified within Stramenopiles (5). Nowadays, Blastocystis spp. is the most commonly reported intestinal protozoa with many subtypes indistinguishable microscopically (6). Blastocystis spp. has strict diversity based on molecular identification and to date 17 subtypes detected. Blastocystis spp. isolated from humans and animals can be assigned as 9 genotypes that are distributed worldwide (7, 8). Molecular epidemiological studies can be useful in clarifying transmission modes, host specificity, treatment algorithms, and resistance to some medications. A significant correlation between certain subtypes and pathogenicity remains to be established. (9). Reported prevalence varies between developed and developing countries and areas. Published studies available in main databases reported frequencies of 5% and higher. In some studies, all collected samples were positive for Blastocystis spp. (10-12). In most studies, the dominant human subtype is 3, and other less common subtypes have different prevalence in different countries e.g., the main subtype in China and Germany is subtype 2, in Japan, it is subtype 6, and in England is subtype 4 (13, 14). In Iran, different studies have detected various frequencies and subtypes. Although molecular diagnostic studies carried out for the genotype identification of Blastocystis spp. from humans in Iran have reported the ST1, ST2, ST3, ST4, ST5, ST6, and ST7, the most common genotype was ST3 (15, 16).
The frequency and genotype identification of Blastocystis spp. in Bandar Abbas, Hormozgan province, South of Iran has not been determined. This cross-sectional study was conducted to obtain data about the Frequency of Blastocystis spp. and determine molecular epidemiology and genotype distribution.
Ethics Considerations
The study was conducted in accordance with the ethical guidelines of the Declaration of Helsinki and protocols reviewed and approved by the Ethics Committee of Hormozgan University of Medical Sciences IR.HUMS.REC.1401.017.
Direct Examination
This descriptive cross-sectional study was conducted in 2022 on 378 human stool samples collected from the patients referred to Shahid Mohammadi Hospital and the city health center. Samples were analyzed by direct and molecular methods. Stool samples from the city health center and Hospital were transferred to the laboratory of the parasitology department of the Faculty of Medicine in a special sampling box. Direct smears and formalin ethyl acetate (concentration) were performed to detect positive samples for Blastocystis spp. parasites. A part of each sample was used for direct smear with saline and Lugel's stain.
DNA Extraction
About 200 mg of stool was used for DNA extraction. A stool DNA Extraction kit (100 prep. FPKT001.0100. Kiagene, Iran) was implemented to extract genomic DNA from stool specimens according to the manufacturer’s instructions. To ensure the accuracy of the method, quality, and quantity of DNA in samples, all microtubes were checked via NanoDrop device.
Primer Design
To design the primer, previous studies were reviewed and the primers of these studies were analyzed (17). Using the following primers (Forward primer: 5'- GGA ATC CTC TTA GAG GGA CAC TAT ACA T-3', Reverse primer: 5' – TTA CTA AAA TCC AAA GTG TTC ATC GGA C- 3'), fragment of 590 bp for Blastocystis spp. identified. Primers were based on a small subunit of ribosomal DNA (SSU rDNA).
Polymerase Chain Reaction (PCR)
DNA samples were stored at -20˚C until use. To amplify segments of approximately 590 bp fragment, a PCR reaction was performed in the final volume of 20 μL. Each reaction contained 10 μL of Master Mix, 0.5 μL of primer F, 0.5 Primer R, 5 μL sterile distilled water, and 4 μL extracted DNA. The cycle algorithm was designed as an initial activation step at 95˚C for 1 min, followed by 35 cycles at 95˚C for 1 min, 35 cycles for annealing at 60 C for 1min, 35 cycles at 72˚C for 2 min, with a final extension step at 72˚C for 7 min. All PCR reactions were done by the BIO-RAD thermocycler system (C1000 Touch; Bio-Rad Laboratories Inc., Hercules, CA, USA).
Gel Electrophoresis
The PCR products were electrophoresed on 1.5% agarose and after optimal conditions stained with Gel red (Biotium Cat. 41003). Gel analyzed by UV transillumination system (Cambridge, UK).
Sequencing of Positive Samples and Phylogenetic Analysis
Sanger dideoxy sequencing technology was used to determine the sequences of positive samples for Blastocystis spp. The Sanger sequencing method is a highly sensitive way to determine the order of nucleotides in an organism (18). All positive samples of the study were sequenced and subtypes were identified. Phylogenetic analysis was performed using the MEGA X software with the Neighbor-joining (NJ) method. The phylogenetic tree’s reliability was assessed by the maximum likelihood method with one thousand bootstrap replications (19, 20). Blastocystis lapemi was used as an outgroup for the Phylogenetic tree.
Statistical Analysis
Statistical data were analyzed using the chi-square test. In all cases, P-values below 0.05 were considered as significant. All statistical analyses were performed using the Statistical Program for Social Sciences (SPSS) version 26 (SPSS Inc., Chicago, Ill., USA).
The population study consisted of patients referred to Bandar Abbas Hospital and medical enter. A total of 378 cases were enrolled in a cross-sectional study in 2022. By direct examination, 14 cases (3.7%) were positive for Blastocystis spp., and 8 cases (2.2%) were approved by PCR technique. Four cases of Giardia were identified and 5 patients were positive for Entamoeba coli.
Phylogenetic Analysis and Genotype Identification
Molecular methods revealed that the 8 positive samples (Figure 1) for Blastocystis spp. were classified as ST1 (4 cases), ST3 (3 cases), and ST2 (1 case) (Figure 2). Statistical analysis showed that there are no significant differences between infection and age, gender, residency location, and education level (P<0.005).
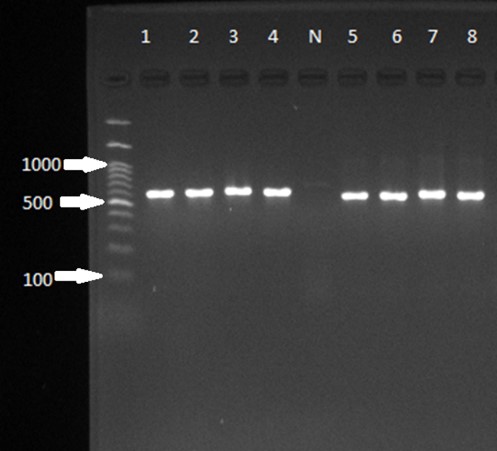
Figure 1. Gel electrophoresis of the 18S rRNA gene for Blastocystis spp. PCR amplicons for genotyping. The ladder, positive samples, and size of bands are visible on the gel. The negative sample is distinguishable.
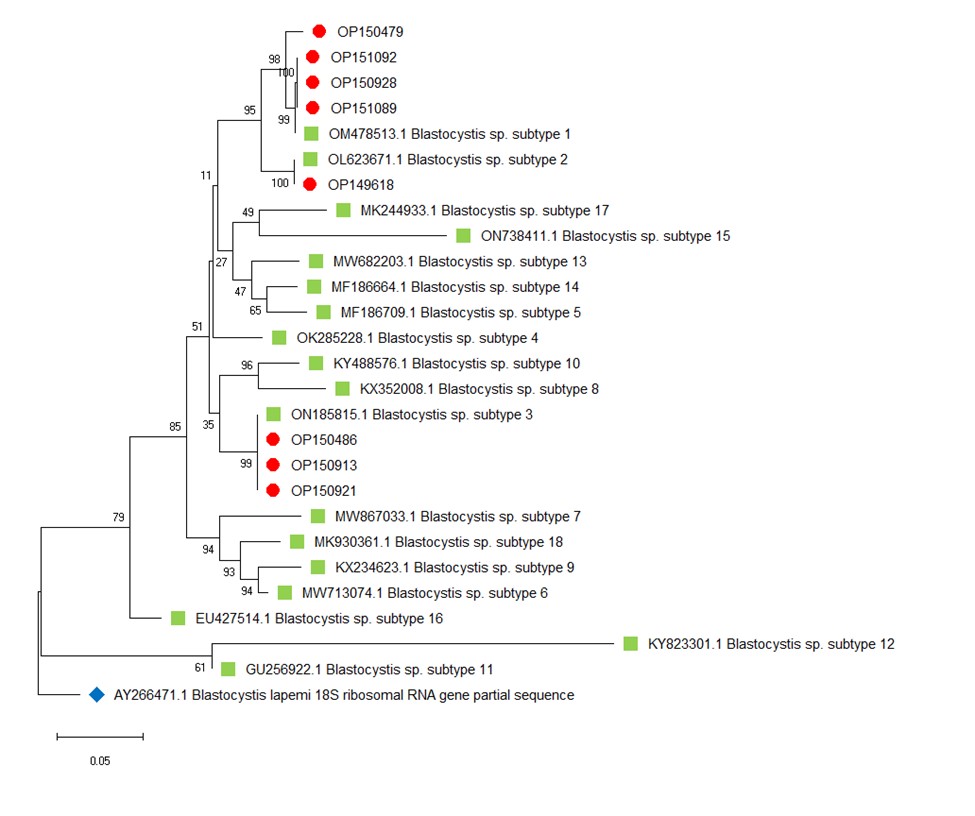
Figure 2. Phylogenetic analysis for Blastocystis spp. Genotypes.
Notes: The phylogenetic tree’s reliability was assessed by the maximum likelihood method with one thousand bootstrap replications and the evolutionary distances were analyzed using the Kimura 2-parameter method. Blastocystis lapemi was used as an outgroup for the Phylogenetic tree.
Nucleotide Sequence Accession Number
The GenBank accession numbers of the 18S rRNA gene of Blastocystis spp. determined in this study are OP149618, OP150479, OP150486, OP150913, OP150921, OP150928, OP151089, and OP151092.
Blastocystis spp. shows genetic diversity in patients and according to identified genotypes, signs, mode of infection, and treatment algorithms could be distinct and different (21). To determine Blastocystis subtypes in Bandar Abbas, Hormozgan province, South of Iran, we designed a survey on stool samples of referred patients to hospital and clinical centers of Bandar Abbas city. Our findings demonstrated that the most common genotype is ST1. Maleki et al. conducted a study in 2022 on 950 stool samples by culture method and conventional PCR. The study isolated ST1-ST6 and ST3 was the dominant genotype (22). They concluded that microscopic examination can't diagnose all positive samples accurately, which is in agreement with our study that we have discrepancies between positive samples by direct and PCR methods (14 positive, 8 positive). Salehi in 2017 investigated samples of the patient in northern Iran and reported ST3 as the dominant genotype. They isolated ST1,2,3 and ST7 (23). Shaker in 2017 carried out a study on 420 stool samples and detected ST3 as the most prevalent subtype of Blastocystis. This study is similar to a survey of subtypes in Lorestan (west of Iran) (24). Khademvatan and colleagues performed a study in southern of Iran in 2014 and isolated five subtypes that ST3 was frequent. In our study, ST1 was dominant after ST3 (25). Khodabakhsh in 2018 studied 1118 stool samples in Kashan, Iran, and findings demonstrate that ST3 and ST1 are the most frequent isolates. Infection rate was correlated to age and education level (26).
A study from Ahvaz showed that of 481 samples collected, 14.35% were positive for Blastocystis spp. and the frequency of Blastocystis spp. in females and males was 13.36% and 15.26% respectively. There was not any statistically significant (P=0.3) (27). In line with this research, our study results showed that there are no significant differences between infection and age, gender, residency location, and education level (P<0.005).
In our study, 14 patients tested positive by direct microscopic examination, and after the PCR test 8 cases were approved. Molecular tests were repeated and results were similar at all times. Caterine Potes Morales (2020) performed a study on fecal samples and identified ST1 as the dominant genotype which is in agreement with our findings. They demonstrated that some patients that were positive in direct smear, were detected as negative by PCR (28). Sari et al. investigated a study on ore school children's stool samples. The genotypes of ST1, 3, and 4 were diagnosed and ST1 was most dominant. This study compared molecular and culture methods. Results showed agreement between methods (29).
Results of a study from Khuzestan province, southwest of Iran, showed that among 268 stool samples, most isolates were related to ST3 with 29 (56.8%) positive cases and ST1 with 11 (21.6%) cases (30), which were in agreement with our results.
Tamas Suli et al. designed and carried out a study on pig stool samples that investigated the molecular epidemiology of Blastocystis spp. using direct smear, xenic culture, and PCR. They compared the sensitivity of 3 methods in isolation of parasite and concluded that molecular technique can be a choice method to approve Blastocystis spp. and has higher sensitivity. Culture is more suitable than direct stool examination (31). Our findings showed that the frequency of Blastocystis spp. is less than in similar studies and direct examination can’t be the only method for diagnosis of this parasite.
This research was conducted at the same time as the COVID-19 pandemic, and there has been an increase in the observance of personal and social hygiene, as well as a decrease in the use of fast food among different classes of society. Therefore, the frequency of Blastocystis spp. parasite in Bandar Abbas compared to other studies is low. Consumption of anti-parasitic drugs and antibiotics during the pandemic may be one of the reasons for reducing the spread of parasites. It is suggested that other investigations be done after the end of the pandemic.
It is better to survey large regions in southern Iran to elucidate the exact rate of genotype distribution. Our study was designed as an MSc dissertation and had some limitations including time and cost.
Not applicable.
We would like to thank and appreciate all colleagues in the faculties and laboratories of Hormozgan Medical Sciences University for their cooperation in sampling and processing of samples and providing facilities.
This research did no
The GenBank accession numbers of the 18S rRNA gene of Blastocystis spp. determined in this study are OP149618, OP150479, OP150486, OP150913, OP150921, OP150928, OP151089, and OP151092 and can be accessed in GenBank (GenBank: https://www.ncbi.nlm.nih.gov/). All data generated or analyzed during this study are included in this article.
This project was approved and funded by Hormozgan University of Medical Sciences (HUMS) as an MSc dissertation.
Competing interests
The authors whose names are listed in the manuscript certify that they have NO affiliations with or involvement in any organization or financial matters that are considered in this study.
Received: 2023/04/24 | Accepted: 2023/08/1 | ePublished: 2023/09/27
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |





