BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-2031-en.html
2- Research Centre for Cellular Genomics and Cancer Research, Sree Balaji Medical College and hospital, Chennai, India
3- Department of Microbiology, Sree Balaji Medical College and hospital, Chennai, India
Antimicrobial resistance is a major threat to the globe mainly due to the lack of treatment options. In the initial period, the gram-positive organism Staphylococcus spp showed resistance to the drugs but now the gram-negative bacteria Enterobacteriaceae is showing multi-drug resistant to various classes of antibiotics and threatening the medical field (1). These organisms transmit the resistant gene very rapidly through plasmids and transposons therefore; the dissemination of resistance against antibiotics develops very rapidly. Development of resistance in microorganisms is mainly based on three mechanisms including β-lactamases, efflux pumps, and mutation in the bacterial gene which alters the protein's expression and function. A combination of these mechanisms with the inclusion of biofilm formation makes the organism more resistant to the therapeutic agents (2). Antibiotic drugs that possess β-lactam rings are called β-lactam antibiotics. Commonly four groups of β-lactam antibiotics are available namely Penicillin, Cephalosporin, Cephamycin, and Carbapenems. Carbapenem antibiotics also possess a β-lactam ring similar to that of penicillin but with few modifications including the substitution of methylene group in the place of C1 instead of sulfur and the addition of two carbon bonds between C2 and C3 (3).
Carbapenem drugs classified under the broad spectrum antibiotics can destroy both gram-positive and gram-negative bacteria and it is one of the effective drugs used as last-line agents. These drugs inhibit the cell wall synthesis of the bacteria. Though some of the drugs namely ertapenem and meropenem show low minimum inhibitory concentration (MIC) in Pseudomonas aeruginosa and Acinetobacter baumanii, other drugs effectively work against gram-negative bacteria. These resistant organisms produce the β-lactamases which inactivate the antibiotics. Carbapenem drugs are widely used to treat the multidrug-resistant organism (4). The increase in Carbapenem-resistant is a challenging scenario in the medical field because these organisms escape from conventional treatment. Because of the increased resistance of microorganisms 50% of the bloodstream infection results in the mortality of the patients. Continuous increase in the resistance of microorganisms limits the therapeutic option (5).
β-lactamases are the enzymes produced by the bacteria to destroy the antibiotics by hydrolyzing the β-lactam amide. Various classes of β-lactamases enzymes are produced by the microorganism to hydrolyze the drug. New Delhi Metallo-β-lactamase (NDM) was classified under the class B family of metallo- β-lactamase family which contains various metals like zinc as a cofactor. Organisms possessing this gene can destroy various carbapenem antibiotics including meropenem, imipenem, doripenem, and ertapenem (6). In the present study prevalence of β-lactam bla-NDM in Klebsiella spp isolated from the various clinical samples was observed.
Collection of Microorganism
In this study 100 isolates of Klebsiella spp were collected from the different samples collected from central diagnostic laboratory, Sree Balaji Medical College and Hospital, Chennai, India from Jan to June 2022. The ethical clearance was obtained from the Institutional Ethical Committee (001/SBMCH/IHEC/2021/1173). To differentiate the organisms at the species level, standard biochemical tests were carried out.
Antibiotic Susceptibility Test
Based on the Clinical and Laboratory Standards Institute (CLSI) guidelines, the antibiotic susceptibility test was carried out by Kirby–Bauer disc diffusion method. Standard antibiotics including, cotrimazole (25 μg) gentamycin (10 μg), cefazolin (30 μg), cefepime (30 μg), imipenem (10 μg), meropenem (10 μg), amikacin (30μg), aztreonam (30 μg), ceftazidime (30 μg), tetracycline (30μg), ciprofloxacin (30 μg), levofloxacin (5μg) was procured from Himedia was used in this study.
Screening of Carbapenem-resistant Organism using Rapidec Carba NP Method
This test was carried out as per the kit instruction (Rapidec Carba NP). In the test strips, 2 mL of the test organism was inoculated and incubated for a few minutes. In the positive carbapenem producer, the color change from red to yellow and orange was observed in the strip after 30 minutes. If the organism is not a carbapenem producer, the red color remains the same.
Detection of the bla-NDM Gene in the Clinical Isolates
The overnight cultured organism was used for the DNA isolation. DNA isolation was carried out using the Qiagen kit. The presence of the bla-NDM gene was screened using the conventional PCR method (Table 1).
Table 1. The bla-NDM gene primer and PCR condition
| Primer | Conditions | Product size |
| 5’-GGGCAGTCGCTTCCAACGGT-3’ 5’-GTAGTGCTCAGTGTCGGCAT-3’ |
94°c for 5 min, 95°c for 30s, 60°c for 30s, 72°c for 30s for 30 cycles, 72°c for 5 min. | 475 bp. |
The genus Klebsiella belongs to the Enterobacteriaceae family and is the common reason to cause nosocomial infections and increases the mortality and morbidity of the patients. This organism quickly acquires the resistant genes via horizontal gene transfer from the plasmids (7). Most nosocomial infections are mainly caused by K. pneumonia and to a lesser extent by K.oxytoca. Studies showed that K. pneumonia inhabited in the environment shares similar biochemical patterns, virulence, and pathogenicity, but the clinical strains show high resistance to the antibiotics which is not observed in the case of environmental strains (8).
In the present study, the prevalence of Klebsiella spp in the tertiary care center and the presence of the blaNDM gene in the isolates were studied. Among the 95 isolates, 80 isolates were K. pneumonia and 15 isolates were K. oxytoca. To differentiate the K. pneumonia and K. oxytoca indole test was performed. K. oxytoca can able to produce indole using the amino acid tryptophan and thus it is indole positive whereas K. pneumonia was indole negative (9).
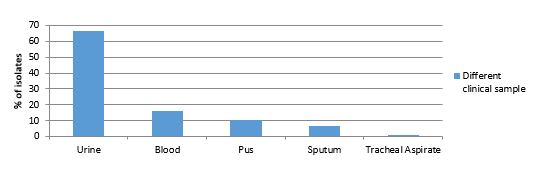
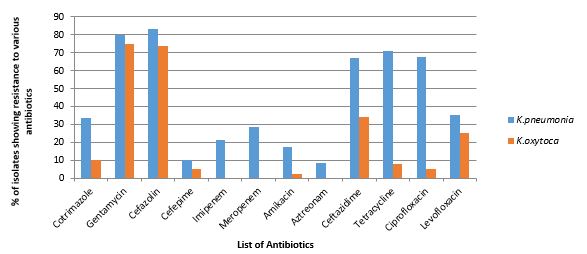
Most of the isolates were from urine (66.31%), followed by blood (15.78%), pus (10.52%), sputum (6.31%), and tracheal aspirate (1.1%) (Figure 1). Most of the isolates collected for the present study showed resistance to gentamycin (80%), cefazolin (82%), tetracycline (72%), imipenem (22%), and meropenem (29%) (Figure 2).
Figure 1. Distribution of Klebsiella spp in different clinical isolates.
Figure 2. Antibiotic-resistant pattern of Klebsiella spp. using different antibiotics.
Isolates showed the least resistance to aztreonam (8%), Cefepime (18%), and Amikacin (20%). Similar to the present study, Nirwati et al showed that Klebsiella isolates were resistant to the first line of antibiotics like gentamycin, tetracycline, etc. (10). Klebsiella isolates collected in 2010 were susceptible to Carbapenem drugs like imipenem and meropenem but the isolates collected during the period of 2015 showed resistant to carbapenem drugs (11). The increase in prevalence of carbapenem drug shown by Klebsiella spp is worrisome. Development of resistance among the organism varies from country to country and it may differ in the same country.
All the isolates were screened for Carbapenem-resistant organisms using the Carba NP method (Figure 3). Among the 80 isolates of K. pneumonia, 45% were Carbapenem producers and 20% of K. oxytoca showed Carbapenem producers. The isolates showed resistance to carbapenem drugs and demonstrated positive for the Carba NP method. An increase in the carbapenemase-producing bacteria is observed in the Enterobacteriaceae family and the increased prevalence was observed in Klebsiella spp. (12).

Figure 3. Showing Carbapenamase-producing organism using Carba NP method. The change in the red to yellow color shows the presence of carbapenem carbapenem-producing organism.
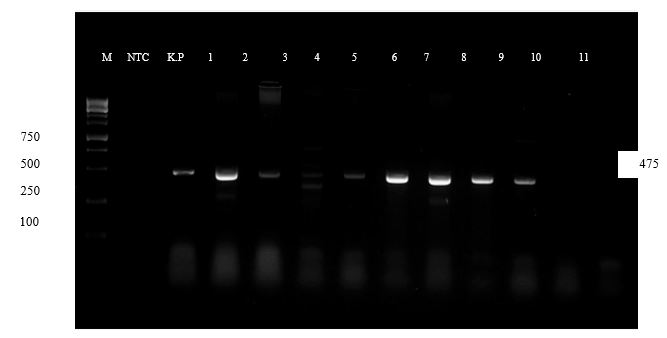
The development of resistance to antibiotics mainly depends on the presence of resistant genes present in the bacteria. The blaNDM gene for New Delhi metallo β lactamase (NDM) strains can break down using carbapenem drugs. The NDM gene is mostly present in the plasmids and these plasmids will enter into other bacteria and transfer the resistant gene to other bacteria (13). The blaNDM gene possesses the novel amino acid close to the active site, which can bind to the β-lactam groups of antibiotics (14). In the present study, among the 95 samples, 20 samples showed the presence of the blaNDM gene (Figure 4). In 19 (23.75%) isolates of K. pneumonia and one isolate (6.6%) of K. oxytoca showed the presence of the blaNDM gene.
Figure 4. shows the presence of the blaNDM gene in the collected clinical isolates
All 20 samples showed the presence of the blaNDM gene that were resistant to meropenem and imipenem drugs. Resistance to imipenem drugs by carbapenem-producing organisms was observed in previous studies conducted in various countries. In Egypt, 52.2% of organisms showed resistance to the imipenem drug and in Greece, all the isolates collected from the hospital showed resistance to the imipenem drug (15). In the present study, 18% of the isolates showed the development of resistance to aztreonam. Usually, the presence of the blaNDM gene showed resistance to most of the β lactam drugs but not to aztreonam. In the present study, the resistance to aztreonam drugs was observed. It might be due to the development of other mechanisms of resistance including efflux pumps or the higher production of Cephalosporinase (16).
The presence of the blaNDM gene in K. pneumonia resulting in resistance to various antibiotics is worrisome. Detection of other carbapenem-producing genes will help study the mechanism of antibiotic resistance which is the limitation of this study. This study suggests that using combinatorial drugs helps in the recovery of the patient because, for diagnostic laboratories, it is not easy to screen for the presence of genes in all the clinical isolates.
This study showed the prevalence of the bla NDM gene in K. pneumonia clinical isolates. The prevalence of the blaNDM gene in the carbapenem-resistant organism was observed in 24% of the samples. Prescription of rational antibiotic therapy can be followed to prevent the spread of antimicrobial resistance.
The authors want to acknowledge the Central diagnostic laboratory, Sree Balaji Medical College and Hospital for providing the samples for the study. The authors also want to acknowledge the research & and development wing of Sree Balaji Medical College for providing the facilities for doing PCR study.
Institutional Ethical Committee reference number: 001/SBMCH/IHEC/2021/1173.
Conflicts of Interest
The authors declare no conflict of interest.
M. R.: Experimental studies, Manuscript preparation, data collection.
M.K.K.: Experimental Design, Experimental studies, Manuscript preparation& editing.
Ch. S.: Experimental Design, Manuscript review, and final decision of the manuscript
N.N.: Experimental design, data collection.
Received: 2023/03/13 | Accepted: 2023/07/27 | ePublished: 2023/09/27
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |