BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1963-en.html
2- Department of Biology, Faculty of Basic Sciences, Rasht Branch, Islamic Azad University, Rasht, Iran ,
Extensive use of antibiotics is associated with increased antibiotic resistance. Antibiotics are used in medicine, veterinary medicine and in animal husbandry as growth promoters that lead to increased resistance among bacteria. The increase and spread of antibiotic-resistant bacterial pathogens threaten the effective treatment of infectious diseases. Resistance factors are often associated with mobile genetic elements such as transposons and conjugated plasmids and thereby are transferred among bacteria (1-3).
Enterococcus spp. is not only an important member of the normal flora of gastrointestinal tract in human and animal but also have clinical importance in causing infections, the ability to create resistance to several antibiotics and their high capacity to receive genetic components through the transfer of plasmids or transposons and propagate resistance genes to other species. These makes this group of bacteria a suitable group for ecological research of antibiotic resistance encoding genes (4-7). On the other hand, these bacteria are able to survive for months in different harsh environmental conditions including low pH and tolerate a broad range of temperatures. So widely distributed in their environment and easily transmitted via the fecal-oral route (8, 9).
Many antimicrobial agents found in livestock belong to those used to treat human enterococcal infections (such as ampicillin, gentamicin, and virginiamycin). For example, the spread of vancomycin-resistant enterococci is possibly linked to using Glycopeptides as growth stimulants in animal and poultry nutrition (10). Enterococcus spp. are intrinsically resistant to several antimicrobials, such as cephalosporins and trimethoprim-sulfamethoxazole, and exhibit low-level resistance to aminoglycosides and β-lactams. Clinical and animal source Enterococcus isolates can also easily acquire and transmit resistant genes and exhibit multi-drug resistance, high level aminoglycoside resistance (HLAR), vancomycin resistance and beta lactamase producing phenotype (11-14).
Like other Gram-positive bacteria, the most common mode of HLAR in Enterococci is attributed to the acquisition of aminoglycoside-modifying genes encoding for aminoglycoside-adenyl transferase, -acetyltransferase and -phosphoryle transferase. Clinically, the bifunctional gene aac(6′)-Ie-aph(2″)-Ia mediates resistance to almost all aminoglycosides except streptomycin and is associated with high-level gentamicin and kanamycin resistance with minimum inhibitory concentration (MIC) values >500 μg/mL. The aph(2'')-Ib gene confers resistance to gentamicin and other types of aminoglycoside. Other resistance-conferring genes found in Gram-positive organisms including Enterococcus spp. are kanamycin modifying aph(3′)IIIa, streptomycin modifying aad(6)Ia and ant(6)Ia (15-17).
Since antibiotic resistance in livestock can threaten human health, the rise and spread of antimicrobial resistance in Enterococcus sp. from domestic and other farm animals has been a concern in recent decades worldwide (16-22). However, in contrast to the large amounts of information on antimicrobial resistance in food- animals, relatively few studies have documented resistance to antimicrobials in bacteria originating from horses. Because of frequent use of similar antibiotics in the treatment of humans and horses, equine origin antibiotic-resistant microorganisms including antimicrobial resistant enterococci, not only affect their health and limits treatment options but also can be treated safety of people who get in contact with these animals and their environment (23-25). Regarding the risk related to the spread of antimicrobial resistance via domestic animals, as the first study in Iran characterizing enterococci from horses, the present research was aimed to investigate the frequency of antimicrobial resistance, high-level aminoglycoside resistance and aminoglycoside resistance associated genes among Enterococcus spp. recovered from healthy Caspian horses in northern Iran.
Equine Study Population and Demographic Data
In one year period, 2021-2022, 140 fresh fecal samples were collected from apparently healthy and non-hospitalized Caspian horses, without antibiotic treatment within the last six months, living in Guilan Province, Northern Iran.
Enterococcus spp. Isolation and Antimicrobial Susceptibility Testing
For isolation of Enterococcus spp., all individual samples were performed in Bile esculin azide agar mediums and were examined according to several bacteriological and biochemical tests including gram staining, catalase tests, bacitracin sensitivity, trimethoprim/sulfamethoxazole sensitivity, growth on 6.5 percent salt medium, growth at 42 ° C, hydrolysis of L-pyrrolidonyl-beta-naphthylamide (PYR) and one Enterococcus spp. isolate per sample was selected and confirmed as Enterococcus using genus-specific primer in PCR reaction as described previously (22).
The disk diffusion assay was used to determine the antimicrobial susceptibility patterns of each Enterococcus spp. isolates according to Clinical and Laboratory Standards Institute (CLSI, 2020) recommendations. Using Penicillin, Ampicillin, Ciprofloxacin, Levofloxacin, Erythromycin, Tetracycline, Gentamicin, Streptomycin, kanamycin and Amikacin (Padtan Teb, Iran), resistance phenotype of the samples was determined. Resistance to vancomycin was determined by determining the minimum inhibitory concentration by broth microdilution method.
Detection of high-level aminoglycoside resistant (HLAR) isolates
For this reason, the minimum inhibitory concentration (MIC) of gentamycin and streptomycin was determined by broth microdilution method based on CLSI guidelines (CLSI, 2020).
Biofilm Forming Assay
Biofilm forming assay was performed in 96-well microtiter plate as described previously (22). with some modifications. In brief, standard overnight cultures (1.5×108 CFU/mL) of test bacteria were diluted 100 folds in Tryptic soy broth containing 1% glucose and 200 µL of each culture dilution was transferred into individual wells of a flat-bottomed polystyrene microtiter plate. After incubation at 37°C for 24 hours, the wells were rinsed three times with PBS to remove planktonic bacteria and subsequently fixed by methanol for 20 min. The wells were stained with 200μl of 0.02% crystal violet and rinsed with distilled water for 5 minutes. Biofilm was quantitatively analyzed by adding 200μl of 33% glacial acetic acid to each well after drying the plates and reading their OD at 492nm by ELISA reader. Strains were evaluated as strong (OD> 4 × ODc), moderate (4 × ODc> OD> 2 × ODc), weak 2 × ODc> OD> ODc), and negative biofilm (OD ODc) based on optical absorption. Staphylococcus epidermidis ATCC 35984 strains and Staphylococcus epidermidis ATCC 12228 strains were used as positive and negative biofilm formation control respectively.
DNA extraction and PCR screening for aminoglycoside resistance genes
Isolates were cultured in Brain Heart Infusion (BHI) broth at 37 °C for 48h. DNA extraction was then performed using the DNA extraction kit of gram-positive bacteria of Cinnagen Company (Cat. No. PR88161) was used.
The aminoglycoside resistance-related genes including aac(6')-Ie–aph(2'')-Ia, aph(2'')-Ib, aph(3′)-IIIa and ant(6)-Ia were evaluated in aminoglycoside resistant isolates, using previously reported primers and PCR conditions (Table 1). The reaction was performed in 25 μl, including 2 U of Taq polymerase, 2.5 μL 10X buffer, 10 mM dNTPs, 20 pmol of each primer, 4 mM MgCl2 and 5 μL of template DNA. Thermocycler thermal treatment was initially denatured at 95°C for 4min, 30 thermal cycles with denaturation at 94°C for the 45s, annealing according to Table 1 for 30s, extension at 72°C for 50s, and final extension at 72°C for 10 minutes. PCR products were detected by electrophoresis using a 1% agarose gel and confirmed by sequencing.
Table 1. Oligonucleotide primers used in this study
| Reference | Amplicon size (bp) | Annealing Tem. | Sequences 5' to 3' | Primer |
| (22) | 112 | 57 | TACTGACAAACCATTCATGATG | Enterococcus spp.-F |
| AACTTCGTCACCAACGCGAAC | Enterococcus spp.-R | |||
| (22) | 220 | 55 | CCAAGAGCAATAAGGGCATA | aac(6')-Ie–aph(2'')-Ia-F |
| CACTATCATACCACTACCG | aac(6')-Ie–aph(2'')-Ia-R | |||
| (22) | 597 | 55 | CACTATCATACCACTACCG ACTGGCTTAATCAATTTGGG |
ant(6)Ia -F ant(6)Ia -R |
| (17) (17) |
958 | 56 | GCAAATGGCACAGTATAATATGC | aph(2'')-Ib-F |
| GCTTGTGTTTGTAGCAATTCAG | aph(2'')-Ib-R | |||
| 663 | 56 | GATACGGAAGGAATGTCTCC | aph(3′)IIIa -F | |
| GCTTGATCCCCAGTAAGTC | aph(3′)IIIa -R |
Statistical analysis
The correlation between biofilm forming ability and the frequency of aminoglycoside resistance genes in test bacteria was analyzed using SPSS software and the Chi-square test. P£0.05 was considered significant.
Enterococcus spp. Isolation and Antimicrobial Susceptibility Testing
Overall, 100 Enterococcus spp. were isolated from 140 horse fecal samples (71.43% recovery). Resistance of isolates to selected antibiotics was detected as; Vancomycin 6%, Penicillin 95%, Ampicillin 89%, Ciprofloxacin 24%, Levofloxacin 16%, Erythromycin 31%, Tetracycline 10%, Gentamicin 32%, Streptomycin 68%, Kanamycin 55% and Amikacin 58%. Among them, 72 isolates were resistant to at least one aminoglycoside and 43% of the isolates demonstrated multiple antibiotic resistance (MDR) phenotypes. Vancomycin was the most potent and Penicillin was the less efficient antibiotic.
Detection of HLAR isolates
Among 100 tested isolates, 38 HLAR Enterococci including 12 high-level gentamicin resistant (HLGR) and 26 high-level streptomycin resistant (HLSR) were determined in tested isolates (MIC values >500 μg/mL).
Biofilm Forming Ability
Among 100 Enterococcus isolates, 67 (67%) were able to form biofilms. Among the positive isolates, around half (34 isolates) were weak biofilm-former. Moderate and strong biofilm formation was observed in 20 and 13 isolates, respectively.
Frequency of aminoglycoside resistance genes
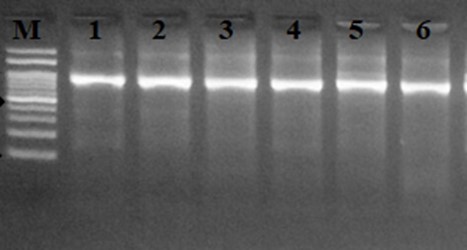
The frequency of 4 aminoglycoside resistance associated genes was investigated among 72 Enterococcus spp. which were resistant to at least one aminoglycoside antibiotic. Bifunctional aminoglycoside resistance associated gene, aac(6′)-Ie-aph(2″)-Ia, was the most common gene and detected in 73.61% (53/72) isolates. The frequency of aph (3´)-IIIa, aph(2'')-Ib and ant(6)Ia genes was 69.44%, 48.61% and 70.83% respectively, among aminoglycoside resistant isolates. All of gentamicin resistant isolates harbored aac(6′)-Ie-aph(2″)-Ia and aph(2'')-Ib genes. The results of agarose gel electrophoresis of PCR products of investigated genes are presented in Figures 1-4.
Figure 1. Agarose gel electrophoresis of aac (6´)-aph (2´´) with approximate size of 220 bp.
Figure 2. Agarose gel electrophoresis of aph (3´)-IIIa with approximate size of 663 bp.
 |
 |
| Figure 3. Agarose gel electrophoresis of aph(2'')-Ib with approximate size of 958 bp. | Figure 4. Agarose gel electrophoresis of ant (6)Ia with an approximate size of 597 bp. |
Correlation between biofilm formation and frequency of resistance genes
Except for vancomycin, resistance to selected antibiotics and the frequencies of all tested aminoglycoside resistance genes were significantly higher in biofilm-positive isolates (P<0.05). All strong biofilm-former isolates harbored at least three aminoglycoside resistance genes.
Over the last years, enterococci have emerged as major nosocomial pathogens with the increasing prevalence of antimicrobial resistance, representing an increasingly important public health problem (7). The acquisition and accumulation of resistance genes in these bacteria lead to the selection of MDR strains including high level resistance to aminoglycosides and an increase of MIC for ampicillin that removed the opportunity of taking advantage of the synergism between these two groups of antimicrobial agents for therapy (26).
In the present study, a total of 100 Enterococcus spp. was isolated from fresh fecal samples of apparently healthy horses living on private farms around the Rasht city. Antibacterial susceptibility testing revealed high-level resistance to penicillin (95%) and ampicillin (89%). Also, 72% of isolates were resistant to at least one tested aminoglycoside and 38% of isolates showed high-level gentamicin and/or streptomycin resistance phenotype. The high frequency of resistance to beta lactams and aminoglycosides may be correlated with the wide use of these antibiotics in veterinary medicine in Iran. Furthermore, our study revealed the presence of multiple aminoglycoside resistance genes in horse fecal enterococci. The frequency of aac(6′)-Ie-aph(2″)-Ia was 73.61% and kanamycin resistance gene aph (3´)-IIIa was detected in 69.44% of aminoglycoside resistant isolates. The occurrence of these genes could be concerned in human health. aph (3´)-IIIa was not detected in 5 kanamycin resistant isolates. This result suggests other resistance mechanisms including the role of the other resistance genes or mechanisms for this phenotype. Resistance to gentamycin is a potent indicator of resistance to other aminoglycosides but not for streptomycin (14). We detected high frequency of streptomycin resistance and high frequency of ant (6)-Ia, that code for resistance to streptomycin (70.83%) in test isolates. Although we didn’t find ant (6)-Ia in 17 streptomycin resistant enterococci but these strains contain other resistance genes including aph (3 ') – IIIa which plays a major role in the emergence of resistance to streptomycin as described previously (27).
Frequency of HLAR and aminoglycoside modifying genes in Enterococcus spp. isolated from humans, animals and the environment have been described in different studies (20, 22, 27-30).
In accordance with our results, high level aminoglycoside resistance was detected in 47.1% of Enterococcus spp. collected from feces of healthy animals and urine of dogs, previously (7). Also, high frequency of resistance associated genes in aminoglycoside resistant isolates were reported by Klibi et al., (2015). These researches detected the ant (6)-Ia and aac(6′)-aph(2″) genes in all aminoglycoside-resistant and aph(3′)-IIIa gene in all kanamycin-resistant enterococci isolated from faecal samples of food animals in Tunisia (17). In another study, aac(6′)Ie-aph(2″)Ia and aph(3′)IIIa were detected in 63.37 and 16.83 Enterococcus spp. isolated from farm animals (13). Among environmental Enterococcus spp isolates, Zaheer et al. (13) detected the ant (6)-Ia and aph(3′)-IIIa at lower frequencies in E. faecalis from wastewater (25%, 42%), as well as in clinical E. faecalis (29%, and 26%) and E. faecium (40%, and 43%). These variations in the frequency of resistance genes in different studies can be related to the types of samples and the treatment strategies in different geographical regions. In contrast with our results, HLG and HLS detection rate were low (1 an 3 isolates respectively) among horse enterococcal isolates and aac(6')Ie-aph(2'')Ia and ant(6)-Ia genes responsible for HLG and HLS were not detected in equine origin resistant isolates in a study conducted by Kim et al. in Korea (22). These data indicate an increasing prevalence of aminoglycosides resistance in equine origin enterococci. The emergence of high frequency of resistant strains in existing work may be explained by the increasing transfer of aminoglycoside resistance genes through plasmids and transposons, frequent availability and the excessive use of these antibiotics without a veterinary prescription. However, recently published data indicate high prevalence of antibiotic resistance in enterococci isolated from water
resources in Guilan province (8), our research is the first report of the frequency of antibiotic and aminoglycoside resistant associated genes in enterococci isolated from livestock sources in northern Iran.
Our findings demonstrate the presence of multi aminoglycoside resistance encoding genes in Enterococcus spp., isolated from equine normal microflora in northern Iran. Since enterococci can acquire and transfer genes horizontally, may exchange resistance genes with human pathogens. So, spreading these isolates in the environment can be a great threat to public health. These data support the ongoing concern about proper monitoring of antimicrobial use in veterinary. In this way, preventing overuse of antibiotics, preventing prolonged exposure of animals to low doses of antibiotics and adherence to prescribed veterinary antibacterial treatments are recommended.
The authors thank the Islamic Azad University, Rasht Branch, for their support.
Elham Nouri was involved in collecting samples, conducting experiments, and writing the manuscript. Study design, analyzing the results, and correcting of the manuscript were done by Leila Asadpour.
The authors didn't receive any financial support.
Conflicts of Interest
The authors declared no conflict of interest.
Received: 2023/01/16 | Accepted: 2023/04/26 | ePublished: 2023/06/26
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |










