BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1679-en.html


 , Nastaran Saadat2
, Nastaran Saadat2 

 , Shahab Mahmoudvand3
, Shahab Mahmoudvand3 

 , Hossein Vakilimofrad4
, Hossein Vakilimofrad4 

 , Salman Khazaei5
, Salman Khazaei5 

 , Nastaran Ansari3
, Nastaran Ansari3 

 , Razieh Amini6
, Razieh Amini6 

 , Farid Azizi Jalilian7
, Farid Azizi Jalilian7 


2- Department of Virology, Faculty of Medicine, Kerman University of Medical Sciences, Kerman, Iran
3- Department of Medical Virology, Faculty of Medicine, Hamadan University of Medical Sciences, Hamadan, Iran
4- Department of Medical Library and Information Sciences, School of Paramedicine, Hamadan University of Medical Sciences, Hamadan, Iran
5- Research Center for Health Sciences, Hamadan University of Medical Sciences, Hamadan, Iran
6- Molecular Medicine Research Center, Hamadan University of Medical Sciences, Hamadan, Iran
7- Department of Medical Virology, Faculty of Medicine, Hamadan University of Medical Sciences, Hamadan, Iran ,
In December 2019, human-to-human transmission of respiratory illness (pneumonia) with fever and sputum was discovered in the respiratory tract. The main factor of this disease is a kind of virus called Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). The virus belongs to Coronaviride, which has the largest genome among RNA viruses, and the genome of this non-segmented virus is a single positive-stranded RNA (1) that has four genera: alpha, beta, gamma, and delta (2). SARS-CoV-2 belongs to the β (beta) genus (3). Coronaviruses mainly cause respiratory infections, and other strains, such as severe acute respiratory syndrome (SARS) and Middle East Respiratory Syndrome (MERS), are associated with infection and high mortality in public health (4). Globally, up to now, there have been about 262 million confirmed cases of COVID-19, including about 5.2 million deaths, reported to WHO (5).
SARS-CoV-2 is involved in lower respiratory tract infection, which leads to acute pneumonia, pulmonary edema, acute respiratory distress syndrome, heart failure, kidney damage, especially in elderly patients with underlying disease (diabetes mellitus, hypertension, etc.) (6, 7). The disease can progress rapidly and expose patients to acute respiratory distress syndrome, resulting in death due to cytokine storms and organ failure (8). Because of the high prevalence, mortality, and epidemic of COVID-19, it is necessary to detect it as soon as possible for faster treatment and medical interventions (4, 9). The definitive method of diagnosis is a CT scan of the lungs and sampling with a nasopharyngeal swab for real-time PCR (10, 11). In this disease, proinflammatory cytokines are secreted, and the vascular system is stimulated (12). Acute-phase proteins (APPs) are reactive proteins that manifest directly in the body as part of the stress response. Serum amyloid A (SAA) is a non-specific APPs that increases in the early stages of inflammation and infection and raises in bacterial and viral infections (3, 9, 13). The function of SAA is known as a cytokine-like protein in cell-cell communication and feedback in inflammatory, immunological, neoplastic, and protective pathways (14). SAA can activate various inflammatory cells via binding to HDL (3). Hepatocytes produce the SAA in response to inflammatory cytokines such as TNF-α, IL-1β, and IL-6. In healthy individuals, SAA protein levels are between 0 and 10 mg/L; under inflammatory conditions, it can increase exponentially, even reaching 1000 mg/L or more in some cases (15). As mentioned above, SAA can be used as an early diagnosis of Covid-19 disease.
Early diagnosis can prevent dangerous consequences and drastically reduce treatment and diagnosis costs. In this research, we conducted a systematic review and meta-analysis of all relevant papers to determine whether SAA as an indicator is involved in the early diagnosis of COVID disease.
2.1. Data sources
This review study was performed from the onset of the COVID-19 epidemic until June 2021 based on systematic review articles and meta-analysis (PRISMA) (16). This study was conducted in 5 stages, including subject design and search strategy, screening and collecting articles from investigated databases, review of inclusion and exclusion criteria, qualitative evaluation, and statistical analysis of data. In order to prevent bias in the study, each of the above steps was performed by two researchers independently. Finally, agreement with the results was obtained by the third researcher.
2.2. Search Strategy
To identify related studies, we systematically searched international databases, including PubMed, Scopus, Science Direct, Cochrane, Embase, Web of Science, and Google Scholar search engines, without restriction on time until June 2021. Also, the Reference list of relevant studies was reviewed separately to find all existing articles in this field. This process was conducted by two researchers independently. Due to the insensitivity of internal databases to search operators (such as AND, OR, NOT). The search was done by the relevant keywords including diagnosis, "Serum Amyloid A", "COVID-19", human "SARS Cov 2", COVID-19 "[Mesh]," Serum Amyloid A Protein "[Mesh], COVID-19 "[Mesh], Human Serum Amyloid A, Early Diagnosis, "Respiratory Disease" using conjunctions (such as AND, OR, NOT). Mesh keywords were also used to maximize and make the results more comprehensive.
2.3. Inclusion and Exclusion Criteria
Inclusion criteria were a meta-analysis of original research articles that examined the level of serum amyloid A protein and Covid-19 disease and the relationship between this biomarker and Covid-19 disease. Exclusion criteria also included: 1- Insufficient data such as lack of quantitative reporting of serum amyloid A protein level, 2- Deletion of articles where the full text of the article was not available, letter to editors, review articles, case reports, animal studies, non-scientific articles. It should be noted that there is no limit on the time of publication or the language of the articles.
Based on the treatment procedures and diagnosis of the new coronavirus, the clinical classification of COVID-19 patients was as follows: (A) Mild type: Mild clinical signs, without the usual change in the lungs on CT imaging. (B) Moderate symptoms: Fever, respiratory symptoms, and CT imaging are commonly shown. (C) Severe type: respiratory distress (respiratory rate ≥30 breaths per minute), mean oxygen saturation ≤93 at rest or relative arterial blood oxygen pressure concentration ≤ 300 mm Hg (D) Critical type: Respiratory failure requires mechanical ventilation, shock, or ICU admission for combined organ failure (17). Based on the above classification, in this work, all patients were divided into two groups: 1- non-severe, mild and moderate, and 2- severe, including severe and critical.
2.4. Data Extraction
Based on the above explanations, in the initial search, 230 articles related to the subject were obtained, which after deleting common cases and inhuman research, reached 73 articles. Finally, reviewing the full text of 16 related studies, ten articles were deleted due to a lack of necessary criteria, and six articles were included in the study due to the mentioned conditions. The items extracted from the articles used in this study were the first author's name, year of publication, country, type of study, study population by gender, and the number of control groups (if used control group) shown in Table 1.
2.5. Quality Assessment
Two researchers separately evaluated the articles' quality using the Newcastle-Ottawa Scale (NOS) checklist (17). Furthermore, any disagreement was examined by a third researcher.
2.6. Statistical analysis
Summary standardized mean difference (SMD) was estimated using random-effects meta-analysis. Heterogeneity among the study results was investigated using the chi-square test (2χ), and the amount of heterogeneity was investigated using I2 statistics. Publication bias was examined using Begg and Egger statistical tests (17). The statistical analysis was performed using the Stata 11.0 (Stata-Corp, College Station, TX, USA).
Figure 1 shows the results of the literature review and selection process. A total of 420 potentially relevant articles were identified from the initial literature review (Three hundred ninety-two for major international databases and 28 for hand searching and reference lists of the identified articles). After removing the duplicates, 230 articles remained, and then the authors excluded 152 articles by screening the titles and abstracts. After full-text examination, 72 articles were excluded due to improper study design or missing data. Finally, 6 studies met the eligibility criteria for meta-analysis. The complete characteristics of included studies are shown in Table 1. Three of the six included studies had a case-control design, and the other was a cohort study. All studies are published in English.
Table 1. The characteristic of studies included in the quantitative meta-analysis stage
| Row | Author (ref number) |
Year | Country | Population Male/Female |
Design | Sample size/ Control | Non severe (Mean±SD) mg/l |
severe (Mean±SD) mg/l |
Control (Mean±SD) mg/l |
NOS Score | Quality |
| 1 | Riham Abdel-Hamid Haroun et al. (1) | 2021 | Egypt | 89/61 | C-C | 150 / 50 | 105.57±17.32 | 171.89±51.96 | 11.94±8.69 | Moderate | High |
| 2 | Qian Liu et al. (4) | 2020 | China | 45/39 | C-C | 84 / 30 | 14.70± 41/65 | 65.75±132/015 | 5.50±7/615 | Moderate | High |
| 3 | SL Liu et al. (3) | 2020 | China | 108/117 | C | 225/ 0 | 3.91±63/21 | 48.57±653/161 | No Control | Good | Good |
| 4 | Lu Li et al. (4) | 2020 | China | 39/33 | C-C | 72/20 | 148.94±54.58 | 260.58±42.67 | 3.94 ± 0.87 | fair | Moderate |
| 5 | Huan Li et al. (9) | 2020 | China | 75/57 | C | 132 / 0 | 123.57±75.81 | 171.91±56.89 | No Control | fair | Moderate |
| 6 | Yalan Yu et al. (25) | 2020 | China | Not reported | C | 3265/ 0 | 13.30±1190/1 | 24.74±930/97 | No Control | Moderate | High |
Note: C-C: case-control; C: cohort
NOS Score: Newcastle-Ottawa Scale

Figure 1. The flow diagram of selected articles in meta-analysis.
3.1. Severe and Non-Severe Group
Our results showed that the levels of SAA protein were significantly higher in the severe group than in the non-severe group (SMD=-0.91, 95% CI: 1.18-1.59, -0.23). Results of the Beggs test (P=0.2) and egger test (P=0.05) indicated no evidence of publication bias in the reported SMD (Figure 2).
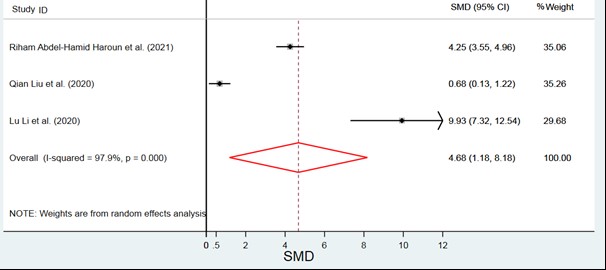
Figure 2.Comparison of severe and non-severe groups. Mean level of serum amyloid A protein in COVID-19 patients based on a random-effects model. The midpoint of each line segment estimates the SMD and line length of the 95% confidence interval in each study. The rhombic mark indicates the mean level of serum amyloid protein A for all studies. The unit is in mg/ml.
3.2. Severe and Control Group
the SAA protein levels in the severe group were significantly higher than the control group (SMD=4.68, 95% CI: 1.18, 8.18). Results of the Beggs test (P=0.6) and egger test (P=0.44) indicate no evidence of publication bias in the reported SMD (Figure 3).
Figure 3. Comparison of severe and Control groups
3.3. Non-Severe and Control Group
No statistical significance was observed between the non-severe and control groups (SMD=3.25, 95% CI: -041, 6.91). Results of the Beggs test (P=0.52) and egger test (P=0.28) indicate no evidence of publication bias in the reported SMD (Figure 4).

Figure 4. Comparison of Non-severe and Control groups
COVID-19 is a rapidly and highly prevalent epidemic with symptoms ranging from mild fever to severe respiratory distress syndrome (ARDS) that complicate diagnosis, prognosis, and treatment (19). Therefore, early diagnosis and appropriate treatment are essential in reducing the complications and mortality of COVID-19 infection. When assessing a patient with COVID-19 infection, biomarkers can help with early diagnosis. Biomarkers are quantitative measurements used clinically for many reflective conditions (1, 19).
Many studies show that certain patients with severe COVID-19 may suffer from cytokine storm syndrome (CSS), which occurs when many WBCs activate and produce inflammatory cytokines (1). TNF-, IL-1, and IL-6 are inflammatory cytokines that cause liver cells to produce SAA, an acute-phase plasma protein (20). In pathological conditions such as viral diseases, SAA is considered better than CRP for timely diagnosis (21). Detection of virus nucleic acid by PCR can confirm its presence or absence. However, it cannot show the progression of the disease, so the need for sensitive indicators that can monitor the onset and progression of the disease is essential. One of these factors detecting the disease in the first stage is inflammatory proteins that are very helpful in preventing the disease process (3). SAA combines rapidly with high-density lipoprotein (HDL) in inflammation. Therefore it can be assumed that people with fatty livers have a higher risk of activating SAA protein than normal people in inflammatory diseases, including SARS-CoV-2 (22). Since the production of this protein takes place in liver cells, if liver function is impaired, its secretion is disrupted during inflammation (9, 22, 23). Therefore, it can be assumed that people with liver diseases such as cirrhosis and hepatitis will face problems in the production process of this protein. Because SAA protein is associated with inflammatory cytokines such as Il-1, Il-6, and TNF, if these cytokines are disrupted, secretion and function of SAA protein will be impaired more than in immunocompromised people and underlying illness, causing a challenge in diagnosing covid-19 (3). In previous research, patients with SARS showed considerably higher levels of SAA, suggesting that SAA might be utilized as a biomarker to track the course of respiratory disorders (24). Therefore, the present study was performed to evaluate the level of SAA as an agent for the early detection of COVID-19 disease. This study's results showed a correlation between SAA level and the severity of COVID-19 infection. Therefore, this biomarker can be used to supplement the early diagnosis of covid-19 disease. Using these early diagnostic factors can increase treatment prognosis and minimize mortality.
In this work, the results of our analysis showed that the SAA level was significantly higher in the severe group than in the non-severe group. Also, the SAA level in the severe group was significantly higher than in the control group. However, no statistical significance was observed between the level of SAA between the non-severe group and the control group.
This study had some limitations, including a. the inability of some information sources to search for combination keywords; b. lack of a single scale to assess the level of amyloid protein A in some published studies; c. failure to provide statistical analysis of amyloid protein A levels in severe and non-severe groups of COVID-19 patients; d. not using the control group in some studies; f. impossibility of accurate and comprehensive analysis due to the limited number of studies.
The critical point is that some asymptomatic patients or cases that can become severe can increase the risk of death due to the severity of the disease and lack of timely diagnosis. In this regard, preventing disease progression from mild to more severe stages can be a promising strategy for reducing disease mortality (25). However, the activity of other biomarkers has not yet been found in this disease, so we urge more research around the world to better understand the changes mentioned in this study.
The result of our work indicates that SAA might potentially be used for the early diagnosis of COVID disease, but more studies are needed.
This study was financially supported by Hamadan University of Medical Sciences (grant no: 36696). Ethics Committee Code is IR.UMSHA.REC.1400.348 and Design number is 140006305301.
None.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Received: 2022/04/24 | Accepted: 2022/10/6 | ePublished: 2023/03/30
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |





