BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1665-en.html
2- Cellular and Molecular Gerash Research Center, Gerash University of Medical Sciences, Gerash, Iran
3- Gerash Al-Zahra Fertility Center, Gerash University of Medical Sciences, Gerash, Iran ,
Ovarian cancer is the seventh most common malignant tumor and the eighth leading cause of cancer death in women worldwide (1). In 2020, 313,959 new cases were diagnosed; unfortunately, 207,252 people died from this disease (2). According to statistics, ovarian cancer has been ranked 24th and 19th death-related disease in Iran and worldwide, respectively (3). Its exact cause is not known. However, a number of risk factors such as having a family history of cancer, stress, smoking, being overweight or obese, drinking alcohol, aging, premature menstruation, late menopause, and undergoing fertility treatment are linked to this type of cancer (4). Microbial infections contribute to around 15 to 20% of all cancer cases. CMV is a ubiquitous herpesvirus that causes lifelong persistence and affects approximately 83% of the world's population (5-7). CMV is a multifaceted beta herpesvirus that usually causes mild discomforts but is fatal in immunocompromised individuals. Several mechanisms are involved in the carcinogenicity of this virus, such as the genome, transcripts, and proteins identified in various cancer types, including colorectal, prostate, breast, and ovarian cancers (8, 9). In Iran, CMV genomes have been detected in breast and colon cancer, oral cavity and larynx carcinoma, and gastric cancers (10, 11). Both BKV and JCV are non-enveloped polyomaviruses that contain a 5Kb circular double-stranded DNA genome and humans are the sole hosts for both viruses (12). These viruses also cause chromosomal deviation in cells and recruit neighboring cells by a 'hit and run' mechanism. They also establish latently on living tissues and their T-Ag is detected in various cancers (13, 14). Several studies have been published regarding the possible correlation between some viral infection and ovarian cancer in Iran (15-18); however, no research have been conducted in Iran and the Middle East to investigate the impact of JCV and BKV, either alone or in conjunction with CMV, in ovarian cancer. Therefore, this study was conducted to determine the frequency and association of CMV, JCV and BKV with malignant, borderline, benign, and normal ovarian tissues in Shiraz, Iran.
In this case-control study, a total of 250 ovarian tissue biopsies were collected from Faghihi Hospital, affiliated with Shiraz University of Medical Sciences. All cases operated from January 2015 to January 2020. An expert pathologist reviewed and confirmed the definite diagnosis of all cases and divided them into 4 groups, including group 1: malignant 93 (37.2%), group 2: borderline 47 (18.8%), group 3: benign 65 (26%), and group 4: control 45 (18%). Figures are available in previously published research (15). The study was approved by the local Ethics Committee of GUMS (GUMS IR.GERUMS.REC.1400.011).
DNA Extraction and Qualification
After deparaffinization of all paraffin-embedded tissues, total DNA was extracted using a commercial kit (Yektatajhiz Inc., Tehran, Iran). The concentration and quality of the extracted DNA were evaluated by nanodrop spectrophotometer and PCR assay for the β -globin gene, respectively (Table 1). Negative samples for amplifying the β-globin gene were excluded from the study. The extracted DNA samples were kept at -20 until the real-time PCR test was used.
Tabe 1. The Primer Sequences and Characteristics Used in This Study
| Locus | Primers | 5' to 3' Sequence | Size,bp | Reference |
| β- globin | PCO3 | 5’ACACAACTGTGTTCACTAGC-3’ | 110 |
(19) |
| PCO4 | 5'CAACTTCATCCACGTTCACC-3' |
Real-time PCR for BKV, JCV, and CMV
Viral DNA was quantified using a real-time PCR method with the BK-JC-Virus (Cat No: BKJC/ISIN/100) and CMV-DNA (Cat No: CMV/ISIN/100) detection kits (Gene Proof, Brno, Czech Republic), according to the manufacturer's instructions. A DNA sequence overlapping the boundary between the genes encoding the VP1 and VP2 proteins of BK- and JC-Virus and the conservative sequence encoding the 4 IE antigen of CMV was the target of the PCR reaction. The amplifications were carried out using the MIC qPCR cycler (BioMolecular System, Australia). The test was repeated two runs to validate the results, and negative control without a template and positive controls were included in each run.
Statistical Analysis
SPSS software version 23 (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. Two statistical tests were used for data analysis, including the t-test and the chi-square test. A P-value below 0.05 was considered statistically significant.
Demographic and pathological results
The histopathology of patients with ovarian cancer is shown in Table 2.
Table2. Histopathology of ovarian cancer patients
| Histopathology | Malignant | Borderline | Bening | Total |
| Serous adenocarcinoma | 68(33.2%) | 33(16.1%) | 43(21%) | 144(70.3%) |
| Mucinous adenocarcinoma | 14(6.8%) | 9(4.4%) | 22(10.7%) | 45(21.9%) |
| Endometrioid adenocarcinoma | 7(3.45%) | 3(1.45%) | 0(0%) | 10(4.9%) |
| Clear Cell carcinoma | 4(1.95%) | 2(0.95%) | 0(0%) | 6(2.9%) |
| Total | 93(45.4%) | 47(22.9%) | 65(31.7%) | 205(100%) |
The mean age of participants was 44.0 ± 11.4 in the age range between 13-83 years. The mean age of participants in the test with malignant, borderline, benign, and normal controls were 55.18±12.4, 44.53±13.3, 40.39±11.7, and 34.2 ± 12.1, respectively.
DNA quality
Because all tissue samples tested positive for the human β-globin gene, the quality of the extracted DNA was deemed adequate.
Real-time PCR for BKV, JCV, and CMV
In this study, none of the borderline, benign ovarian tumor tissues or the normal group was positive for the presence of CMV, JCV, or BKV DNA, but our results showed that one, two, and three samples of malignant tumors were positive for the BKV, JCV, and CMV genome, respectively (20). The frequency of BKV, JCV, and CMV in the malignant group was not significantly different between the four groups of this study (Table 3).
Table 3. The frequency of BKV, JCV, and CMV in four study groups.
| Group |
Malignant tumor (n=93 ) |
Borderline tumor (n=47 ) |
Benign tumor (n= 65 ) |
Normal group (n=45 ) |
P-value |
|||
| Total | Stage I (n=41 ) |
Stage II (n=24 ) |
Stage III (n=28 ) |
|||||
| BKV | 1(1.07%) | 0 (0%) | 1(4.16%) | 0 (0%) |
0 (0%) |
0 (0%) | 0 (0%) | 0.64 |
| JCV | 2(2.15%) | 0 (0%) | 1(4.16%) | 1(3.57%) | 0 (0%) | 0 (0%) | 0 (0%) | 0.34 |
| CMV | 3(3.22%) | 0 (0%) | 1(4.16%) | 2(7.14%) | 0 (0%) | 0 (0%) | 0 (0%) | 0.17 |
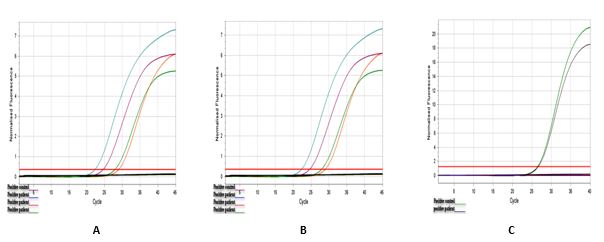
Figure 1. Amplification plot of real-time PCR tests
Detection of Cytomegalovirus DNA in Ovarian Cancer B) Detection of JCV DNA in Ovarian Cancer C) Detection of BKV DNA in Ovarian Cancer
Ovarian cancer is the most common and life-threatening type of female gynecological cancer (21). The exact cause of ovarian cancer remains unidentified; however, a number of studies have detected viral genomes in cancerous ovarian tissue. Consequently, it is presumed that some viral infections may be implicated in ovarian cancer (22, 23). These viruses cause modulation and tumor-promoting inflammation, which may play a fundamental role in developing quantized, asymptomatic cancer (24, 25). Worldwide, rare studies report controversy over the role of BK, JC, and CMV infections in ovarian cancer. Also, there is no data from the Middle East. The present study evaluated the presence of BK, JC, and CMV genome in cancerous and benign ovarian tissue to shed light on viral infections that may play a role in carcinogenesis.
In our study, BKV, JCV, and CMV's DNA fragments were not detected in any of the borderline, benign, or control lesions. Only 1 BKV, 2 JCV, and 3 CMV out of the 93 malignant samples demonstrated the presence of the viruses.
Although, subsequently, statistical analysis showed no association between CMV, JCV, and BKV infection and ovarian cancer, the low prevalence of these viral infections in our investigation could be interpreted in different ways: 1) First of all, there may be no connection between the investigated viral agents and ovarian cancer, 2) The low prevalence of these viral agents does not rule out their involvement in this process; it could also be due to hit-and-run oncogenesis mechanisms, in which the virus initiates the oncogenesis process but then clears from the host, 3) Finally, some oncogenic agents act indirectly by inducing chronic inflammation that can promote or initiate cancer.
The result of this study coincided with studies that failed to detect BK, JC, and CMV infection in ovarian carcinoma tissue. In 2010, Idahl et al., investigated the presence of the BK and JC viruses in tissue samples, but none of them were detected in the ovarian cancer samples nor the benign controls (26). However, Ingerslev et al. found CMV DNA in only 0.5% of their patients, so they did not support an association between CMV and ovarian cancer (27).
In contrast, Paradowska et al. demonstrated that 70% of cancerous ovarian tissues contained CMV DNA, which was significantly higher than in benign tumors (28).
In another study by Shanmughapriya et al., CMV infection was detected in 50% of cases with ovarian cancer and 50% of borderline ovarian tumor cases, whereas it was not detected in the normal control group (29).
In a study by Randstad and colleagues also found CMV IgM in 12% of ovarian cancer patients and 3% of patients with benign tumors, although it was absent in controls (30).
As previously stated, there is a disagreement study results, which may be due to the patient genetic background, environmental factors, sample size, and patient lifestyle (sexuality). Also, several technical issues, such as the detection method (type and sensitivity), different types of sample (fresh or fixed tissues), or even extraction procedures, or the possibility of cross-contamination, could cuse different results.
This study was limited by its small sample size and the lack of availability of some clinical materials.
In conclusion, our study showed a very low frequency of BK, JC, and CMV DNA in ovarian cancer tissues. Regarding controversial issues and limited literature, additional studies with a larger sample size and more sensitive methods are needed to better identify the role of these viruses in ovarian cancer. These investigations may provide new insights into ovarian cancer's pathogenesis and new strategies for using antiviral therapy in oncology patients.
The present study was the result of a research project which was approved by the local Ethics Committee of Gerash University of Medical Sciences (IR.GERUMS.REC.1400.011). The authors wish to thank Jamal Sarvari at the department of Bacteriology and Virology, Shiraz University of Medical Sciences, for their invaluable assistance in this investigation.
All financial support is provided by Gerash University of Medical Sciences (99000033).
Conflicts of Interest
The authors confirm that there are no known conflicts of interest associated with this publication, and there has been no significant financial support for this work that could have influenced its outcome.
Received: 2022/02/8 | Accepted: 2022/06/26 | ePublished: 2022/09/9
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |