BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1505-en.html
2- Brucellosis Research Center, Hamadan University of Medical Sciences, Hamadan, Iran
Alcaligenes genus, is a Gram-negative, aerobic, rod-shaped, non-fermentative bacteria with amphitri-chous flagella and rarely non-motile. Additionally, some strains of Alcaligenes are capable of anaerobic respiration, but they must be in the presence of nitrate or nitrite; otherwise, their metabolism is respiratory and never fermentative; the genus does not use carbohydrates (1).
Strains of Alcaligenes such as A. faecalis and A. xylosoxidans are found mostly in the intestinal tracts of vertebrates, decaying materials, dairy products, water, and soil (2-4); they can be isolated from the human respiratory (2, 3) and gastrointestinal tracts (5) and wounds in hospitalized patients with compro-mised immune systems (6, 7). They are occasionally the cause of opportunistic infections, including nosoc-omial sepsis (2, 8). Alcaligenes sp. is an important hospital pathogen that is morphologically very similar to Pseudomonas aeruginosa (9) and maybe confused with Pseudomonas species (8).
Alcaligenes species causes bacteremia in immune-deficiency patients with situating catheters (10). The bacterium, along with other infections such as P. aeruginosa, has been reported to cause respiratory tract infections in cystic fibrosis patients. At present, the clinical aspects of this bacterium are not very clear in relevant infections (9, 11). Due to high rate of infections caused by this bacterium and its resistance to some common antibiotics, different mortality rates have been observed in patients with infections caused by this bacterium (12).
Routinely phenotypic methods such as dedicated culture media, biochemical tests, and API (Analytical Profile Index) detection kits use to diagnose this bacterium (4). Although phenotypic methods are a precise identification method, molecular methods are more effective and more accurate in identifying non-typical isolates. Already, molecular methods such as PCR, DNA fingerprinting, RAPD-PCR, and PCR assay based on 16S ribosomal DNA, are used to investigate the molecular nature of this bacterium (11, 13, 14).
Antibiotic resistance among pathogenic bacteria especially strains causing nosocomial infections, is particularly important. Also, due to acquisition of antibiotic resistance genes by bacteria over time in different geographical areas and changes in the pattern of bacterial susceptibility to different antibi-otics, choosing the right antibiotic for treatment has become a challenge. (16) Based on the mentioned statements and due to few published data about the accurate diagnosis of clinical strains of Alcaligenes in Iran, in this study as the first time in Iran, we investigated the biochemically and genetically confirmation of the presence of A. faecalis and A. xylosoxidans in clinical samples and their antimicrobial susceptibility patterns.
Isolates tested
In a descriptive-analytical study from September 2019 to March 2020 at a Sina hospital in Hamadan, Iran, we analyzed all 36 isolates collected from hospitalized patients who were phenotypically identified by the referring clinical microbiology laboratories as Alcaligenes. We get the ethical approval letter from the institutional research ethics review committee of Hamadan University of medical sciences. (NO. IR.UMSHA.REC.1396.443).
Phenotypic profiling
The bacteria studied in this research were isolated from blood and urine samples of hospitalized patients in Sina hospital. To phenotypic identification, all isolates were grown on selective media such as SIM, TSI, MR, VP, Citrate, Urea agar, and MacConkey agar (Merck, Germany) and incubated at 37ºc for 24 hours. To confirmation the purity, the samples were examined by Gram staining. Phenotypic characteri-zations were performed on all isolates, using protocols for Gram-negative, non-fermentative bacteria such as motility tests, oxidative fermentation of glucose and lactose, Bile-esculin, DNase, Nitrate reduction, and Indole reactions. Clinical isolates approved by biochemical experiments were stored at -20 °C. Following the phenotypic and biochemical tests, we used the PCR test to more investigate accurately the detection and confirmation of the phenotypic results.
DNA extraction, PCR amplification, and DNA sequencing
Bacterial genomic DNA for PCR amplification was extracted by the boiling method as previously descry-bed (15). The NanoDrop spectrophotometer (Thermo Scientific™ 840274100) was used to determine the amount of extracted DNA. The purity of DNA (OD A260/280) was measured in ng/µl. The quality of the extracted DNA was assessed on 1% Agarose gel.
The PCR was performed in BioRad T100 PCR Thermocycler with AX and 77F-r gene-specific primers, to genetically confirm the phenotypically identified A. xylosoxidans and A. faecalis, respectively. The specific primers used for the amplification of the AX gene were AX-F 5̒̒ GCAGGAAAGAAA CGTCGC GGGT 3̒̒ and AX -R 5̒̒̒ ATTTCCATCTTTCTTTCCG 3̒. The 77F-r specific primers (77F-r-F 5̒ GGCGGACGGGTGAGTAATA 3̒ and 77F-r-R 5̒ CTGCAGATACCGTCAGCAGT 3̒) were also used to confirm A. faecalis (16). The PCR reaction mixture of 12.5 µl contained 1 µl of DNA template (20 ng), 0.5 µl of each primer (concentration of each primer was 0.5 µM), 6 µl of Taq DNA Polymerase Master Mix RED (amplicon, Denmark) and, 4.5 µl distilled water was used. The first PCR step was performed at 95°C for 3 min and was followed by 30 cycles of denaturation (95 °C for 1 min), annealing (55 °C and 58 °C for 1 min in annealing AX and 77F-r, respectively), and extension (72 °C for 1 min). The last step was performed at 72 °C for 5 min. Positive control (A clinical sample confirmed by sequencing) and negative control (Pseudomonas aeruginosa ATCC 27853) were utilized to avoid false-positive results. The amplified PCR products were analyzed using 2% agarose gels and stained with GelRed® nucleic acid stain. An amplified PCR product corresponding to the expected size of AX and 77F-r genes was used for sequencing. Sequences obtained for each of the analyzed genes were assembled and compared to available sequences in Genbank, using the BLAST (Basic Local Alignment Search Tool) algorithm of the NCBI (National Center for Biotech-nology Information) (16).
Antimicrobial susceptibility assay
We performed the antimicrobial susceptibility tests for each isolate by the disc diffusion method (Kirby-Bauer). The results were interpreted as either sensitive, intermediate, or resistant according to the Clinical Laboratory Standards Institute (CLSI-2018) susceptibility breakpoints for non-fermenting gram-negative bacteria (17). Antibiotic discs (Mast (UK)), used for the tests included: ampicillin (AP10 ug), trimethoprim/sulfamethoxazole (TS25 ug), ciproflo-xacin (CIP5 ug), imipenem (IMI10 ug), Gentamicin (GM10 ug), Meropenem (MEM10 ug), Ceftazid-ime-(CAZ30 ug), ceftriaxone(CRO30 ug), piperacillin-/tazo-bactam (PTZ110 ug), ampicillin/sulbactam (SAM20 ug), cefepime (CPM30 ug).
Patient Characteristics
Out of 36 samples collected in this study, 19 (52.8%) samples belonged to women, and 17samples (47.2%) were from men. The patients ’age was between 27 and 95 years, with an average age of 58.2.The patients ’age was between 27 and 95 years, with an average age of 58.2.
Genetic Confirmation of Alcaligenes species
The PCR results revealed that only 16 (44.44%) samples were Alcaligenes species, which included 13 (36.11%) samples as A. xylosoxidans using the AX-specific primers and 3 (8.33%) samples as A. faecalis using the 77F-r-specific primers. A 163 bp and a 391 bp PCR fragment were seen following the electrophoresis, corresponding well with the expected size of a part of AX and 77F-r genes, respectively (Fig. 1 and 2).
We compared the AX and 77F-r gene sequences to the whole genome sequences of the genus of Alcaligenes present in the NCBI database. The alignment results demonstrate a 100% identity with the 16S rRNA genes of the A. xylosoxidans and A. faecalis bacteria. This compliance confirmed the specificity of the PCR products as a positive control.
Antimicrobial Susceptibility
The in vitro susceptibility of 36 Alcaligenes isolates to 11 antimicrobial agents is summarized in Table 1. The most susceptibility (80.55%) among Alcaligenes species was to Cefepime, followed by imipenem, piperacillin-tazobactam, and ceftazidime with a 75% rate. Also, the most resistance (92.3 %) was seen against Cefepime antibiotic in 13 A. xylosoxidans isolates followed by ciprofloxacin (76.92%) and meropenem (38.46%) (Table 2).
Figure 1. Agarose gel electrophoresis (2% agarose) of PCR amplified products using species-specific PCR primer sets (163 bp) in A. xylosoxidans strains. Lane M, 100 bp DNA ladder, lane 1: negative control, lane 2: positive control, lane 3, 4: A. xylosoxidans strains.
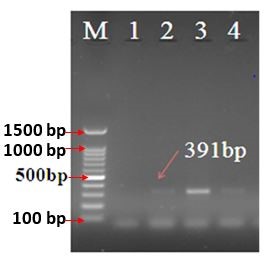
Figure 2. Agarose gel electrophoresis (2% agarose) of PCR amplified products using species-specific PCR primer sets (391 bp) in A. faecalis strains. Lane M, 100 bp DNA ladder, lane 1: negative control, lane 2: positive control, lane 3, 4: A. faecalis strains.
Table 1. In vitro susceptibility profile of 36 Alcaligenes species isolated from hospitalized patients to 11 Antimicrobial agent
| Antibiotic | No. of isolates (%) | ||
| Susceptible | Intermediate | Resistant | |
| Gentamicin | 12 (33.33) | 3 (8.33) | 21 (58.33) |
| Ceftazidime | 27 (75.00) | 1 (2.77) | 8 (22.22) |
| Ceftriaxone | 11 (30.55) | 14 (38.88) | 11 (30.55) |
| Piperacillin/Tazobactam | 27 (75.00) | 5 (13.88) | 4 (11.11) |
| Ampicillin | 9 (25.00) | 6 (16.66) | 21 (58.33) |
| Trimethoprim/sulfamethoxazole | 21 (58.33) | 3 (8.33) | 12 (33.33) |
| Meropenem | 21 (58.33) | 1 (2.77) | 14 (38.88) |
| Ampicillin/Sulbactam | 17 (47.22) | 3 (8.33) | 16 (44.44) |
| Imipenem | 27 (75.00) | 7 (19.44) | 2 (5.55) |
| Ciprofloxacin | 8 (22.22) | 2 (5.55) | 26 (72.22) |
| Cefepime | 29 (80.55) | 1 (2.77) | 6 (16.66) |
Table 2. In vitro susceptibility profile of 13 A. xylosoxidans and 3 A. faecalis strains isolated from hospitalized patients to 11 Antimicrobial agent
| No. of isolates (%) | ||||||
| Antibiotics | Susceptible | Intermediate Susceptibility | Resistant | |||
| A. xylosoxidans | A. faecalis | A. xylosoxidans | A. faecalis | A. xylosoxidans | A. faecalis | |
| Gentamicin | 4 (30.76) | 1 (33.33) | 8 (61.53) | 0 | 1 (7.70) | 2 (66.66) |
| Ceftazidime | 11 (84.61) | 3 (100) | 0 | 0 | 2 (15.38) | 0 |
| Ceftriaxone | 5 (38.46) | 1 (33.33) | 5 (38.46) | 0 | 3 (23.07) | 2 (66.66) |
| Piperacillin/Tazobactam | 10 (76.92) | 3 (100) | 2 (15.38) | 0 | 1 (7.70) | 0 |
| Ampicillin | 7 (53.84) | 0 | 3 (23.07) | 2 (66.66) | 3 (23.07) | 1 (33.33) |
| Trimethoprim/sulfamethoxazole | 11 (84.61) | 2 (66.66) | 1 (7.70) | 0 | 1 (7.70) | 1 (33.33) |
| Meropenem | 8 (61.53) | 3 (100) | 0 | 0 | 5 (38.46) | 0 |
| Ampicillin/Sulbactam | 9 (69.23) | 2 (66.66) | 1 (7.70) | 0 | 3 (23.07) | 1 (33.33) |
| Imipenem | 11 (84.61) | 3 (100) | 0 | 0 | 2 (15.38) | 0 |
| Ciprofloxacin | 2 (15.38) | 1 (33.33) | 1 (7.70) | 1 (33.33) | 10 (76.92) | 1 (33.33) |
| Cefepime | 12 (92.30) | 3 (100) | 0 | 0 | 1 (7.70) | 0 |
Several studies have been conducted to evaluate the immunogenicity of various conjugates, including ALG-TT, ALG-SLNs, PLGA-ETA, ALG-DT, etc. In this study, for the first time, the immunogenic effect of ALG-ETA conjugate was investigated against P. aeruginosa infections.
Our findings showed that antitoxin A antibodies can play a greater protective effect than P. aeruginosa infections. This conjugated compound increases the level of anti-ALG IgG antibodies, and immunization with this type of conjugate can stimulate toxin A-neutralizing antibodies. T
None
This article is an independent study conducted with no organizational financial support.
Conflicts of Interest
The authors declared no conflict of interest.
Received: 2021/10/3 | Accepted: 2022/01/13 | ePublished: 2022/02/10
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |









