BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1449-en.html
2- Department of Microbiology, Ayatollah Amoli Branch, Islamic Azad University, Amol, Iran ,
3- Department of Microbiology, Tonekabon Branch, Islamic Azad University, Tonekabon, Iran
4- Department of Chemistry, Tonekabon Branch, Islamic Azad University, Tonekabon, Iran
5- Department of Genetics, School of Basic Sciences, Tonekabon Branch, Islamic Azad University, Tonekabon, Iran
P. aeruginosa is an opportunistic pathogen that causes infections by exploiting the weakness of the human immune system, especially in patients with severe and immunosuppressive chronic infection (1, 2). The ability to grow at simple nutrition supply and temperatures from 4°C to 42°C, resistance to conditions with high concentrations of salts and dyes, weak antiseptics and many commonly used antibiotics are critical factors involved in achieving its prominence as community-acquired and 11–13.8% of all hospital-acquired infections (2, 3).
Several model system studies demonstrated that many Gram-negative bacteria like P. aeruginosa utilizes types I, II, III, V, and VI secretion systems (TSS) to introduce virulence effector proteins directly into host cells (4). Each TSS is composed of several common secreted factors that play key roles in the pathogenesis of P. aeruginosa strains at various infections. Type III secretion systems (TTSS) is a contact-dependent protein secretion pathway with four exoenzymes effectors, ExoS, ExoT, ExoU, and ExoY, which are associated with acute infections of P. aeruginosa (5, 6). Further investigation showed that one and usually rarely both of the two major exotoxins ExoU and or ExoS express about at all pathogenic strains. In contrast, ExoY and ExoT with minor roles express at most strains. ExoS and ExoU are more common exotoxins in about all invasive and cytotoxic P. aeruginosa strains, respectively (6-9). Exotoxin A (ExoA) is considered the most toxic virulence factor and a potent inhibitor of protein synthesis secreted by P. aeruginosa. Its injection into the extracellular space through the type II secretory system causes inactivation of elongation factor-2 (EF-2), inhibiting protein synthesis and leading to cell death.
Quorum sensing (QS) is a bacterial mechanism of cell-to-cell communication that regulates more than 10% of genes involved in bacterial growth, many virulence factors production, and adaptation to environmental changes (6, 10). QS acts hierarchically through threshold concentrations of small membrane-diffusible molecules called autoinducers (signal molecules). Acyl-homoserine lactones (AHLs), including 3-oxo-dodecanoyl homoserine lactone (3-oxo-C12 HSL) and butyryl homoserine lactone (C4 HSL), are common autoinducers in P. aeruginosa produced by the LasI AHL synthase and RhlI AHL synthase, respectively. Following an increase in bacterial population and diffusion determined concentration of secreted autoinducers into the environment, they subsequently bind to specific transcriptional activators (LasR/RhlR) that trigger expression of genes involved in biofilm formation and coding virulence factors (11).
Recently treatment of P. aeruginosa infections has emerged as a big concern considering a broad range of intrinsic, acquired, and adaptive resistance mechanisms. Therefore, therapies aimed at disrupting the QS signaling cascade and biofilm formation have gradually become a promising alternative to conventional antibiotic therapies. The utilization of nanoparticle (NP) potentials has recently received great attention to overcome the ongoing crises caused by antibiotic resistance (12). Chitosan (CS) and zinc oxide nanoparticles (ZnO-NPs) have been greatly considered promising “green” biocides due to their remarkable biological advantages obtained through nanoscale dimensions. These compounds have been characterized as quorum quenchers (QQ) that disturb bacterial communications resulting in inhibits virulence without interfering with the growth. The mechanism of the antibacterial effect of CS and ZnO on P. aeruginosa by targeting the inhibition of QS and biofilm formation has been reported previously, but gene expression at the mechanism of virulence inhibition following treatment with these NPs being less investigated (13, 14). Therefore, it is essential to understand the effectiveness of nanoparticles on the expression of virulence genes to prevent pathogenesis, particularly at the initial stages. The main objectives of this study were to isolate exoS+, toxA+ and lasI+ strains from burn patients and investigate the inhibitory effect of CS and ZnO-NPs on expression changes of lasI, exoS, and toxA virulence genes in the mid of lag-, exponential- and stationary growth phases of toxA+/exoS+/lasI+ P. aeruginosa originated from burn patients and ATCC27853 standard strain.
Synthesis of CS Nanoparticles
CS-NPs were synthesized by ionic gelation according to Calvo’s method with slight modifications (20). Briefly, a stock solution of chitosan (Sigma, USA) was dissolved in 0.05% (v/v) acetic acid and kept at room temperature under continuous stirring overnight. The pH of the solution was adjusted to 4.7–4.8. Sodium tripolyphosphate (TPP) (Sigma, USA) was dissolved in ultra-pure water to a concentration of 0.5 mg/mL, filtered (0.22 μm, Millipore), and subsequently stirred at 2–4 °C. CS-NPs were formed by adding CS solution dropwise to TPP solution with a volume ratio of 3:1 (CS: TPP) and keeping for 15 minutes at room temperature under magnetic stirring (1000 rpm). Then the obtained solution was centrifuged at 13,000 rpm for 20 minutes, and residual TPP was discarded. The CSNP-containing pellets were freeze dried and kept at 4°C until further experiments.
Synthesis of ZnO Nanoparticles
ZnO-NPs were synthesized by the ultrasonic method using ZnC₄H₆O₄ (Sigma, USA), Sodium Hydroxide (NaOH), and propylene glycol 10% as starting materials (21). For synthesis, 0.3 g of zinc acetate and 0.32 g of NaOH were dissolved in 60 mL of ethanol and exposed to ultrasound for 30 minutes at room temperature. The obtained precipitates were then rinsed with distilled water several times, centrifuged and the remaining solid dried in vacuum desiccators. Finally, different concentrations of ZnO-NPs was resulted by dispersing various concentrations of obtained ZnO in 10% propylene glycol.
Characterization of CS and ZnO Nanoparticles
The morphology and particle size characterization of synthesized NPs were studied using Scanning Electron Microscopy (SEM) (TESCAN, MIRA-3) and Transmission Electron Microscopy (TEM) (Zeis, EM-1030, Germany). Specimens for TEM analysis were dispersed in ethanol and sonicated in an ultrasonic bath for 20 min. The X-ray power diffraction patterns (XRD) of synthesized compounds were recorded on (X’ Pert Pro, Panalytical, UK) with Cu Kα radiation (λ = 1.5412 Å) in the scanning range of 100-800. To examine the characteristic chemical structure of CS and ZnO-NPs, fourier transform infrared (FTIR) spectra was carried out using JASCO INC 410, Japan, in the range of 400–4000 cm−1.
Sample Collection and Isolation of the Bacteria
The samples used in this study were collected from Shahid Zare Hospital of Sari, Iran. In this study, patients suffering from wound infections from 2018-2019 were studied. Samples were collected from burn wounds using sterile swabs and immediately transported in 5 mL of Brain-heart infusion (BHI) broth (Merck, Germany). The BHI tubes were sent to the clinical laboratory and incubated overnight at 37°C. Sub-culture was done on nutrient agar media (Merck, Germany) at 37°C for 24 hours for subsequent identification.
Phenotypic and Molecular Identification of Isolated Bacteria
Preliminary identification of P. aeruginosa has been done with the standard biochemical tests, including Gram stain, catalase, oxidase, oxidation-fermentation (OF) test, the reaction in triple sugar iron (TSI) agar, growth ability at 42°C, Pyocyanin pigment production in Mueller-Hinton agar (MHA) (Merck, Germany) and growth on Cetrimide Agar media (15) on positive cultures. In order to molecular identification, the total DNA of overnight bacterial cultures was obtained using the DNeasy bacterial Mini Kit (QIAGEN, Germany), according to the manufacturer's instructions. The quality of extracted DNAs was evaluated on 1% agarose gel electrophoresis and quantified by NanoDrop™ 2000 system (Thermo Fisher Scientific, USA). PCR amplification of 16S rDNA gene was performed in 25 µL reaction mixture containing 14 µL dd water, 2.5 µL10 × PCR buffer, 0.5 µL of dNTPs (10 mM), 0.75 µL MgCl2 (50 mM), 0.25 µL Taq polymerase enzyme (5 U/µL) (CinnaGen, Iran) 1 µL of each primer (10 pmol) (forward strand primer 5′-GGGGGATCTTCGGACCTCA-3′ and reverse strand primer 5′-TCCTTAGAGTGCCCACCCG-3′), and 5 µL (20 ng/μl) of extracted DNA from P. aeruginosa isolates using the T100 thermal cycler (Bio-Rad). After an initial denaturation at 95°C for 2 min, the reaction mixture was run followed by 30 cycles of 95°C for 20 seconds, 58°C for 20 seconds, and 72°C for 40 seconds, with a final extension at 72°C for five min. The indicated primers were produced by TAG Copenhagen (Denmark) and amplifying a 956 bp product of the 16S rDNA gene of P. aeruginosa (16). P. aeruginosa ATCC 27853 was used as positive control and PCR products were visualized by electrophoresis using a 1% agarose gel stained with ethidium bromide using a UV transilluminator (Kodak Gel Logic 200, USA).
Detection of Virulence Genes by PCR
PCR amplification of lasI, exoS and toxA virulence genes were performed in 25 µL reaction mixture containing14 µL dd water,2.5 µL10 × PCR buffer, 0.5 µL of dNTPs (10 mM), 1 µL of each primer (10 pmol), 0.75 µL MgCl2 (50 mM), 0.25 µL Taq polymerase enzyme (5 U/µL) (CinnaGen, Iran) and 5 µL (20 ng/μL) of extracted DNA from P. aeruginosa isolates. All primers were synthesized by TAG Copenhagen (Denmark) and the sequences of primer sets and amplification conditions are represented in Table 1 and 2. PCR products were separated using 1% agarose gel electrophoresis, followed by visualization of stained gel under UV light.
Quantitative assay of the antimicrobial activity and the effect of sub-MIC of CS and ZnO-NPs on bacterial growth
The determination of minimal inhibitory concentrations (MIC) of CS and ZnO-NPs was performed following the standard broth dilution method (CLSI M07-A8). For this purpose, serial two-fold dilutions of CS and ZnO-NPs in concentrations ranging from 0.125 mg/mL to 8 mg/mL was prepared in Tryptic Soy Broth (TSB) and inoculated with 10 μL P. aeruginosa culture equivalent to a 0.5 McFarland’s standard (108 CFU/mL) and incubated overnight at 37°C. In this study, a medium containing bacterial suspension without NPs and media without bacteria with nanoparticles were used as a positive and negative control, respectively. The lowest concentration of NPs that prevented the visible growth of bacteria was regarded as MIC. One dilution below the MIC was considered the sub-MIC concentration and used for further study (22). For growth curve measurements, diluted overnight clinical P. aeruginosa isolates and ATCC 27853 strain in TSB medium were supplemented with sub-MIC concentrations of CS and ZnO-NPs and incubated at 37°C under continuous agitation for 24 h. The turbidity of grown bacteria was measured at 600 nm using a spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) at the end of each time interval (23). The experiments were performed in triplicate and the results were reported as the mean ± SD (standard deviation).
Table 1. Specific primer sequences targeting lasI, exoS and toxA genes.
| Primer | 5'-sequence-3' | Product size (bp) | Reference |
| lasI | CACATCTGGGAACTCAGC ACGGATCATCATCTTCTCC |
171 | (17) |
| exoS | ATCCTCAGGCGTACATCC ACGACGGCTATCTCTCCAC |
328 | (18) |
| toxA | CACAGGCAACGACGAGGC CCTTGTCGGGGATGCTGG |
710 | (19) |
Table 2. The amplification conditions used for the PCR amplification of lasI, exoS and toxA genes in P. aeruginosa.
| lasI exoS toxA | |||||||||
| Steps | Temperature | Time | Cycle | Temperature | Time | Cycle | Temperature | Time | Cycle |
| Primary denaturation | 95°C | 2 min | 1 | 94°C | 4 min | 1 | 95°C | 5 min | 1 |
| Denaturation | 95°C | 40sec | 30 |
94°C | 1 min | 30 |
95°C | 30 sec | 30 |
| Annealing | 55°C | 1 min | 55°C | 1 min | 58°C | 30 sec | |||
| Extension | 72°C | 2 min | 72°C | 2 min | 72°C | 50 sec | |||
| Final extension | 72°C | 10min | 1 | 72°C | 10min | 1 | 72°C | 10 min | 1 |
RNA Extraction and qRT-PCR anaLysis
qRT-PCR was performed to detect the expression level of QS and virulence-related genes (lasI, exoS and toxA) in lag-, exponential-and stationary- growth phases of P. aeruginosa before and after treatment with CS and ZnO-NPs. Cultivated P. aeruginosa in TSB medium before and after supplemented with sub-MIC concentrations of CS and ZnO-NPs until the middle of each lag-(2 h), exponential (8 h) -and stationary (18 h) growth phases (OD 600; 0.5 McFarland) were collected for RNA extraction. Total RNA was isolated using Hybrid-R™ kit (GeneAll Biotechnology, Korea) based on the manufacturer's instructions and stored at − 80 °C. The quality and quantity of extracted RNAs were evaluated using 1.5% agarose gel electrophoresis and NanoDrop™ 2000 system (Thermo Fisher Scientific, USA), respectively. According to the manufacturer's protocol, extracted RNAs were reverse-transcribed using HyperScript™ RT premix kit (GeneAll Biotechnology, Korea). PCR was performed on a StepOne Plus machine (Applied Biosystems, USA) using a SYBER Green kit (Ampliqon, Denmark) with specific primers listed in Table 3. The reaction was carried out as follows: holding stage at 95°C for 10 min, cycling stage comprising 40 cycles (95°C for 15 s, 60°C for 1 min, 72°C for 30 s), melt curve stage at 60°C for 1 min, and 95°C for 15 s. Beacon designer software was utilized for designing primers based on the deduced complete genome sequence of P. aeruginosa ATCC 27853 from NCBI (National Centre for Biotechnology Information). The rpoD gene was used as an internal control to normalize the expression of the quantified genes. Gene expression levels in CS and ZnO-treated P. aeruginosa was compared to their expression levels in untreated ones using the 2−△△Ct method (24).
Table 3. Primers used in this study for real time PCR of lasI, exoS, txoA and rpoD genes in P. aeruginosa.
| Gene | 5'-sequence-3' | References |
| exoS | TCGGCGTCACTGTGGATG AGAGAGCGAGGTCAGCAGAGTATC |
This study |
| toxA | CGAGCAGGCACAACACCTT CGCACGAGAGCAACGAGAT |
This study |
| lasI | CCGCACATCYGGGAACTCA GATCATCATCTTCTCCACGCCTAC |
This study |
| rpoD | GCTGCTGTCGTCGCTTTCTT AGCATCCTGGCCGACTACAA |
(25) |
Statistical Analysis
Statistical analysis was done using IBM SPSS® Statistics version 19 (IBM® Corp., Armonk, NY, USA). All experiments were performed in triplicate and the data were expressed as mean and standard deviation. The significance of all the statistical tests was determined at P-value< 0.05.
Identification of Clinical Isolates
Twenty-five clinical isolates from one hundred and forty burn samples were identified as P. aeruginosa according to phenotypic and biochemical tests. To confirm detected isolates, the presence of 16S rDNA gene was investigated by PCR amplification. PCR yielded amplicons with predicted sizes of 956bp confirmed the existence of P. aeruginosa DNA in the same twenty-five isolates (Figure 1).
Detection of Virulence Genes
The presence of lasI, exoS and toxA virulence genes in P. aeruginosa were studied by PCR. The detection results (Figure 2) showed that out of 25 P. aeruginosa isolates identified, only 13 (52%) were positive for all the three lasI, exoS, and toxA virulence genes. Positive results with toxA and exoS genes were 25 (100%) and 22 (88%), respectively, while lasI gene was detected only in 14 (56%) of the collected P. aeruginosa isolates.
Figure1. Agarose gel (1%) electrophoresis of PCR products after amplification of 16S rDNA gene (956bp); M: GeneRuler 1000 bp DNA Ladder, Fermentas as size marker; N. 1-5: PCR amplification; N.6: Control negative.

Figure 2. Electrophoresis of PCR products (A) lasI (B) exoS and (C) toxA genes of P. aeruginosa clinical isolates in agarose gel.); M: GeneRuler 1000 bp DNA Ladder, Fermentas as size marker; N. 1-5: PCR amplification; C-: Control negative
Characterization of CS and ZnO Nanoparticles
The structural properties of the CS and ZnO NPs were analyzed using XRD and X’Pert Highscore Plus software. Figure 3 A-D manifests the X-ray diffraction patterns of synthesized CS and ZnO-NPs. The definite line broadening in the XRD patterns of both CS and ZnO indicates that the prepared particles were in the nanoscale range. Diffraction peaks appeared at 2θ = 11.7° and 2θ =19.8° agreed with previous reports (26-28) and can be indexed to the successful synthesis of CS-NPs (Figure 3A). The characteristic of broad peaks with 2θ values of 31.66°, 34.3°, 36.2°, 47.4°, 56.5°, 62.8°, 66.3°, 67.8° and 68.8° corresponding to the hexagonal wurtzite structure of ZnO-NPs confirmed using JCPDS card number 96-900-8878 (Figure 3B) (29). IR characterization of CS and ZnO-NPs were acquired in the range of 400–4000 cm−1, and the spectra are shown in Figure 2. The CS-NPs showed a peak at 3449 cm-1 attributed to stretching vibration of –NH2, –OH groups and the enhanced hydrogen bonding of nanoparticles (Figure 3C). The sharp peaks located at 1658 cm−1 and 1557 cm−1 can be attributed to the formation of –NH3+ and cross-linkage with phosphate groups of TPP. The spectra of CS-NP showed peaks at 1157 and 1073 cm-1 can be attributed to the stretching of the C-O-C bridge and P=O stretching vibration from phosphate groups, respectively (30, 31). The spectral features of ZnO-NPs (Figure 3D) are as follows: absorption at 3391 cm−1 corresponding to the stretching vibrations of the –OH group on the surface of ZnO-NPs, 1641 cm−1 related to asymmetric stretching of zinc carboxylate (C=O) (32), and a signature band at 582 cm−1 as the characteristic stretching mode of Zn-O bond (33-35). The sharp bands assigned to –CO stretching vibration and the existence of metal were found at 946 and 1004 cm−1 (36).
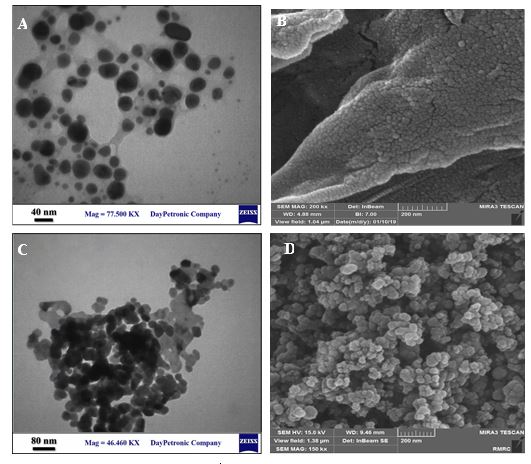
Figure 4 (A, B, C and D) shows the TEM and SEM micrographs of the surface morphological structure of CS and ZnO-NPs. The TEM of CS-NPs (Figure 4A) shows CS-NPs having a spherical shape without agglomeration and an average particle size range of about 20 nm. The SEM image (Figure 4B) also confirmed that the CS-NPs showing spherical structures. The TEM and SEM (Figure 4C and D) images of ZnO-NPs exhibited the ZnO-NPs have grown almost in spherical morphology with an average diameter of about 40 nm. Taken together, good qualities of synthesized NPs were observed from SEM and TEM images.
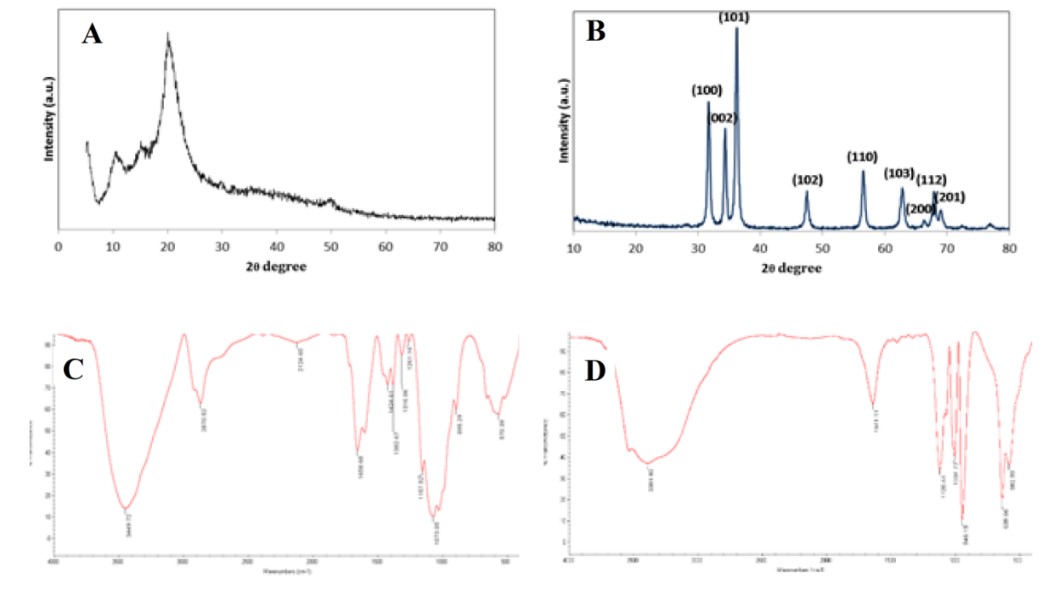
Figure 3. XRD patterns of CS-NPs (A) and ZnO-NPs (B). Fourier-transform infrared spectrum of CS-NPs (C) and ZnO-NPs (D).
Figure 4. TEM of CS-NPs (A), SEM of CS-NPs (B), TEM of ZnO-NPs (C), and SEM of ZnO-NPs
Determination of Antimicrobial Activity of NPs and Growth Inhibition Assay
The MIC of CS and ZnO-NPs have been measured on the 13 P. aeruginosa strains possessed toxA+/exoS+/lasI+ genotype and ATCC 27853 as the standard strain on TSB media containing selected serial concentrations of NPs. In this study, treatment with NPs did not show a similar inhibitory effect at a certain concentration for all the tested clinical isolates. The MIC values of CS and ZnO-NPs were found to be 0.5 mg/mL and 4 mg/mL for five clinical isolates, respectively, while the growth of the rest of the isolates was inhibited at the lower concentrations of mentioned NPs. Therefore, five clinical isolates harboring toxA+/exoS+/lasI+ genotype that showed higher resistance to NPs were selected for further analysis. The MIC values of CS and ZnO-NPs were equal to 0.125 mg/mL and 2 mg/mL for standard strain, respectively (data not shown).
0.25 and 2 mg/mL were used as sub-MIC for clinical isolates and 0.0625 and 1 mg/mL for ATCC 27853 to evaluate their inhibitory effect on growth performance and virulence factors. As a result, ATCC 27853 standard strain showed higher susceptibility to both NPs than clinical isolates in which CS-NP possess higher inhibition activity. The graph shown in Figure 5 depicted the growth inhibition of bacteria supplemented in media containing the respective sub-MIC of NPs. The difference in growth rates of standard and clinical isolates was not statistically significant before and after treatment of CS and ZnO-NPs (P>0.05). Sub-MIC of CS-NPs showed a greater decrease in the growth rate than ZnO-NPs (P>0.05).
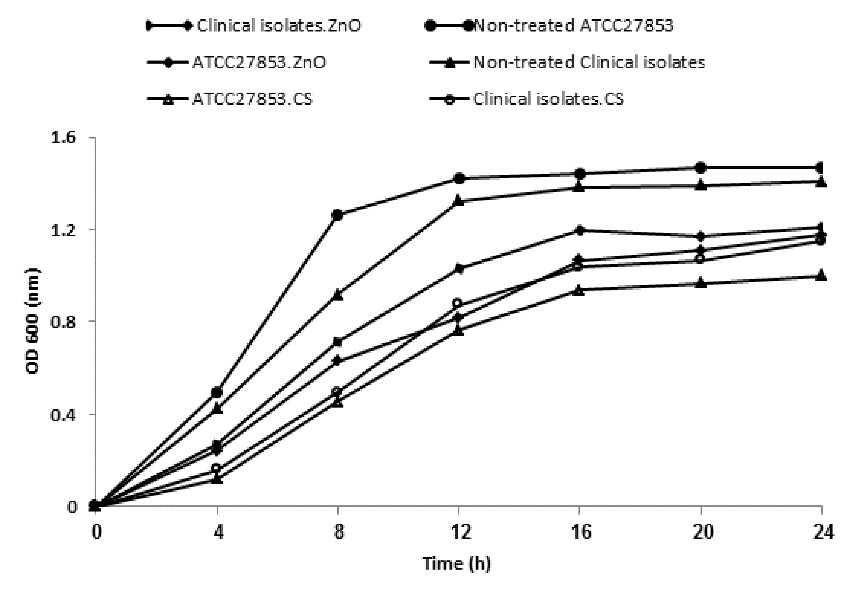
Figure 5. The growth curve of clinical P. aeruginosa isolates and ATCC 27853 before and after treatment with sub-MIC of NPs. Value of the clinical isolates is expressed by the mean of five selected resistance isolates.
Expression of QS and Virulence-related Genes in Response to CS and ZnO-NPs at Different Growth Phases
The relative expression of lasI, exoS, and toxA genes was assessed at the middle of each lag-, exponential-and stationary-growth phases of bacteria in medium supplemented with and without sub-MICs of CS and ZnO-NPs. CS and ZnO-NPs showed a down-regulation with relative expression levels of 0.802- and 0.706-fold change for lasI gene in the exponential growth phase of clinical isolates while in standard strain was increased by 1.21 and 1.07 folds, respectively (Figure 6 A-D). On the contrary, lasI expression level was increased with relative transcriptional levels of 1.048-, 1.037- and 2.72-, 1.67-fold change in lag and stationary phases of clinical isolates treated with CS and ZnO-NPs, respectively, compared to a standard strain (Figure 6 A and D). For CS-NPs, the level of exoS and toxA genes expression in clinical isolates was reduced by 0.699 and 0.962 folds in the exponential growth phase and increased with relative transcriptional levels of 1.15-, 1.194- and 1.131-, 1.02-fold change in lag and stationary phases, respectively (Figure 6 B and C). ZnO-NPs inhibited the expression of exoS and toxA genes in clinical isolates as their expression was decreased by 0.604 and 0.765 folds in the exponential growth phase, respectively (Figure 6 E and F). The fold change in the expression of exoS and toxA genes was 1.53, 1.42, and 1.2, 0.9-fold in lag and stationary phases of clinical isolates treated with ZnO-NPs, respectively. For the standard strain treated with ZnO-NPs, expression of exoS gene showed upregulation during all phases while expression of toxA gene was downregulated except for lag phase (1.43-fold) (Figure 6 E and F).
Figure 6. The relative expression of lasI, exoS and toxA genes in clinical p. aeruginosa isolates and ATCC 27853 after treatment with CS-NPs (A, B and C) and ZnO-NPs (D, E and F) at lag-, exponential-and stationary-growth phases. The data of three replicates shown as the means ± SD (value of the clinical isolates is expressed by mean of five selected resistance isolates). Significant differences between clinical p. aeruginosa isolates and ATCC 27853 were determined by t-test; * P< 0.05, ** P< 0.01, ***P< 0.001.
Understanding the inhibitory effect of NPs on the expression of virulence dependent genes at different growth phases of P. aeruginosa can be an effective alternative approach to antibacterial therapies and overcome infection through interrupting and/or blocking expression of QS-regulated virulence factors (36).
Emerging of multi-drug resistant (MDR) P. aeruginosa strains due to their innate and acquired resistance mechanisms and excessive and indiscriminate use of antibiotics have led to difficult treatment, high morbidity, and mortality, especially in burn-infected patients (37).
To confirm the evaluation of CS and ZnO-NPs effects on the expression of P. aeruginosa virulence factors, the presence of lasI, exoS and toxA genes was first investigated by PCR and isolates containing all three genes were selected for qRT-PCR analysis. In our study, the presence of toxA, exoS and lasI genes was detected in 100%, 88% and 56% of all the burn clinical isolates, respectively. In studies by Yousefi-Avarvand et al. and Amirmozafari et al. the prevalence of toxA among the clinical isolates of P. aeruginosa was reported 90.4%, 97% and 66.7%, 59.8% for exoS, respectively (4, 10).
Nikbin et al. reported the frequency of toxA and exoS genes for 100 clinical isolates from burn-infected patients to be 97% and 67% (38). Our findings showed a high prevalence of virulence-related markers in which 52% of isolates harboring all the three lasI, exoS and toxA that was almost in line with the findings of other studies from various parts of Iran.
Similar to other results, increased frequency of isolates possessing both toxA and exoS toxin genes in this study may confirm their possible contributory role associated with causing infection and emerging resistance strains in burn patients (4). In this study, 13 clinical isolates harboring toxA+/exoS+/lasI+ genotype were subjected to further study. NPs and their nanocomposites especially combine with antibiotics due to their many enhanced properties and excellent biological activities being considered novel nanotherapeutic approaches to overcome infections caused by multidrug-resistant organisms (39). Therefore, the present study was conducted to evaluate the effects of CS and ZnO -NPs on growth performance, QS and virulence-related gene expression at different growth phases in P. aeruginosa.
According to the XRD and FTIR results, all characteristic absorption and diffraction patterns indicated purity and the crystalline structures of obtained CS and ZnO-NPs, similar to previously reported literature (34, 35). TEM and SEM micrographs indicated spherical structure, network formation and agglomeration of synthesized CS and ZnO-NPs that confirmed with the results obtained from XRD.
CS and ZnO-NPs as organic- and inorganic-based nanomaterials are currently studied among the most widely used antimicrobial agents against a wide spectrum of bacterial species as well as P. aeruginosa compared to their counterparts. Nanoparticle-dependent antibacterial mechanisms directly correlate with particle size and concentration, where smaller size and higher concentrations of nanoparticles displayed better antimicrobial activity (40). According to literature reports, P. aeruginosa strains are susceptible to CS-NPs with MIC values ranging from 128 to 4096 μg/mL (41), indicating that the MIC values of 0.125 and 0.5 mg/mL obtained in our studies for standard and clinical isolates are favorable due to the range in which they are. The values of 1600-3200 µg/mL(42), 375 μg/mL (43), 128 µg/mL and 300 mM (44) has been previously reported as MIC of ZnO-NPs for P. aeruginosa depending on the size, stability, and concentration of agent as well as MIC determination methods and growth medium (45). In our study, NPs exhibited concentration-dependent antimicrobial activity against identified isolates with toxA+/exoS+/lasI+ genotypes in which at five isolates, the growth was inhibited at higher concentrations of NPs. ZnO-NPs prevented the growth of tested standard strain and five clinical isolates at concentrations of 2 mg/mL and 4 mg/mL, respectively. Our finding is rather close to those reported by C. de Souza et al. (46) (up to 2 mg/ mL) for ATCC 27853 standard strain. In case of clinical isolates exception for the value reported by Saleh et al. (47), other reported values were lower than the MIC value observed in the present work. In agreement with other studies (42, 48), both NPs showed a lower activity towards the clinical isolates. CS-NPs were able to inhibit the growth of P. aeruginosa at a lower concentration than ZnO-NPs.
lasI, exoS and toxA are major QS and virulence-related genes involved in the P. aeruginosa pathogenesis (8). Several publications have been well studied the potential of ZnO and CS-NPs to prevent virulence factor production and biofilm formation through various mechanisms against P. aeruginosa (14, 36, 49). We further study the effect of synthesized ZnO (40 nm) and CS (20 nm) NPs on the expression level of lasI, exoS and toxA virulence-related genes of five P. aeruginosa (toxA+/exoS+/lasI+) originated from burn patients compare to ATCC 27853 standard stain. To the best of our knowledge, this is the first study to evaluate the expression of mentioned genes with an emphasis on lag-, exponential and stationary-growth phases of P. aeruginosa from burn patients.
In the present study, lasI, exoS, and toxA genes were downregulated due to CS and ZnO-NPs treatment in the exponential phase of clinical isolates with the higher inhibitory effect for CS compared to ZnO-NPs. Similarly, it was proved by Badawy et al. (36) that CS-NP down-regulated the expression of lasI. A similar study by Singh et al. (49), Rao et al. (48), and Saleh et al. (47) supported our results, where ZnO -NPs showed an inhibitory effect on lasI expression level. Furthermore, in ATCC 27853, except for toxA, lasI and exoS genes showed upregulation at the exponential phase. On the other hand, these genes showed upregulation during the lag and stationary phases in clinical isolates. A similar gene expression pattern was observed for toxA in treated CS and ZnO toxA+/exoS+/lasI+ isolates and ATCC 27853 during all growth phases. Our results showed that sub-MIC concentration of CS and ZnO-NPs could decrease the expression of QS-mediated virulence factors only during the exponential phase compared to the other growth phases in clinical isolates. The sub-MIC concentration of CS and ZnO-NPs exhibited similar expression changes for toxA, lasI, and exoS genes during lag, exponential and stationary phases of toxA+/exoS+/lasI+ P. aeruginosa isolates compared to ATCC 27853. In the current study, although the sub-MIC concentration of CS was lower than ZnO-NPs, its inhibitory effect on expression level was more than those induced that by ZnO-NPs. This is due to the positively charged and small size of CS-NPs.
In conclusion, this study is the first to examine the inhibitory effects of CS and ZnO-NPs on the expression of lasI, exoS and toxA virulence genes during lag-, exponential- and stationary-growth phases in P. aeruginosa. The synthesized CS and ZnO-NPs successfully downregulated QS and virulence-related gene expression in the exponential growth phase relative to lag and stationary phase. Gene expression patterns of lasI, exoS and toxA were varied between toxA+/exoS+/lasI+ P. aeruginosa isolates and ATCC 27853 at different growth phases. The higher inhibitory effect of CS-NPs on lasI, exoS and toxA gene expression levels was observed compared to ZnO-NPs. The results of this study can be useful for a better understanding of the inhibitory effects of NPs at overcoming P. aeruginosa harboring lasI, exoS and toxA virulence genes.
Thesis advisors Masood Ghane and Hami Kabossi are acknowledged and appreciated extremely in the research process as well as all employees of the Microbiology Research Laboratory ward in the Islamic Azad University of Tonekabon Branch.
The study protocol approval was provided by the Ethics Committee of Islamic Azad University of Tonekabon (IR.IAU.TON.REC.1399.014).
Mozhgan Pourjafari Collected the data and wrote the paper. Hami Kaboosi conceived and planned the experiments. Masood Ghane Conceived and designed the analysis and collected the data. Babak Sadeghi, Abolhasan Rezaei verified and performed the analysis.
Conflicts of Interest
The authors do not report any financial or personal connections with other persons or organizations, which might negatively affect the contents of this publication and/or claim authorship rights to this publication.
Received: 2021/06/25 | Accepted: 2021/08/10 | ePublished: 2021/09/5
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |