BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1407-en.html

 , Ramak Yahya Raeyat2
, Ramak Yahya Raeyat2 
 , Mohammadreza Mehrabi3
, Mohammadreza Mehrabi3 

 , Taghi Zahraiee Salehi1
, Taghi Zahraiee Salehi1 

 , Jalil Mehrzad Salakojani1
, Jalil Mehrzad Salakojani1 


2- Department of Microbiology and Immunology, Faculty of Veterinary Medicine, University of Tehran, Tehran, Iran ,
3- Department of laboratory Medicine, Faculty of Medicine, Borujerd Branch, Islamic Azad University, Borujerd, Iran
The genus Salmonella consists of rod-shaped, Gram-negative, non-spore-forming, and mostly motile intestinal bacteria with 0.7-1.5 µm in diameter and 2-5 µm in length. These facultative anaerobic bacteria use organic substrates and redox reactions to obtain energy. Most Salmonella serovars produce hydrogen sulfide (H2S) and are not able to ferment lactose, which can easily be detected by growing in an environment containing iron sulfate. There are more than 2,500 salmonella serotypes (serovar) that are classified based on cell wall antigens (O antigen), flagellar antigens (H antigen), and surface or envelope antigens (1, 2).
Salmonella is a very successful intestinal pathogen because it has different strategies to deal with the host immune system. Gastrointestinal mucosa, the first innate immune barrier, is a thick layer of mucus that covers the surface of the intestinal epithelium. Other barriers include macrophage cells, which are innate immune-specific phagocytic cells that belong to one of the cell types present in connective tissues. As mobile and fixed cells, they have important effects on the body's innate immune system (5). Macrophages are classified as mononuclear phagocytic cells, which in addition to macrophages, also include monocytes, promonocytes, and their progenitor cells. The main function of these cells is to "cleanse" the blood, lymph, and other tissues by swallowing or phagocytosis of various particles. Macrophages are taken from the bone marrow, distributed throughout the body, and have different shapes and characteristics, often depending on the tissue in which they are found, the degree of differentiation, and the age or life span of the organism in which they are found (Figure 1) (6).
Over the past few years, the analysis of changes in mRNA expression has been explored in many studies. The results showed significant mRNAs expression changes in many infections, such as Salmonellosis. These expression profiles can be used in the treat-ment, prevention, and diagnosis of infected patients (7, 8). Clinical results obtained from gene expression profiles in different studies have led to different results, which has made their certainty and reprod-ucibility questionable. Of course, it is clear that these differences may be due to the variety of test designs, including patient types, small tissue size, and lack of control samples in the experiments. Although studies of analytical tools have led to these differences and dissatisfaction with the use of these methods, studies still give us good information (9-11).
Classically or alternatively activated macrophages are immune-affecting cells and produce large volumes of lymphokine that are associated with the expression of genes such as IFN-γ, TNF-α, and TNF-β. During the immune response to various infectious agents, M1 and M2 macrophages are activated (12). In immune regulation mechanisms, the polarization of this popu-lation forms the basis of homeostasis. One way to confirm polarization is to express the IFN -γ and TNF -α cytokines for phenotype M1 and TGFβ for pheno-type M2. The aim of this study was to investigate the effect of Salmonella typhi on the activity of human blood macrophage-like monocytes in vitro. For this purpose, the expression of IFN -γ and TNF -α cytokines are used to confirm the polarization of Salmonella typhi-stimulated macrophages (13-16).

Figure 1. Schematic representation of immune cell interactions in Salmonella typhi infection (14).
This cross-sectional descriptive study was perfor-med with ethics code IR.lums.REC.1399.023 in 2021 on 60 samples of feces collected from patients with clinical signs of gastroenteritis admitted to the internal wards of Ayatollah Boroujerdi, Dr. Chamran, Imam Khomeini, and Kosar hospitals, Boroujerd, Iran. The volunteers were given brief explanations by a specia-list and written consent was obtained. A questionnaire was completed to collect patients' demographic information. In this study, people who did not sign a written consent, who had taken antibiotics before sampling, and also had a suppressed immune system were excluded from the study.
Sample Collection
Stool samples were collected in a clean, wide-mouthed plastic container with a tight, leak-free lid. The container was free of preservatives, detergents, metal ions, or toilet paper. At least one gram of stool with normal consistency (about the size of a hazelnut) or 5 mL of diarrhea stool was used for culture. Stool samples were taken fresh and cultured up to 2 hours after sampling.
Isolation and Identification of Salmonella sp.
To identify Salmonella sp., biochemical tests, including oxidase, nitrate, glucose fermentation, and gas production, TSI (H2S +), lactose-sucrose-ONPG, IMViCC (- + -) were performed. The Salmonella typhi ATCC14028 was used as a control purchased from Iran Scientific-Industrial Research Center (17).
Isolation of Peripheral Blood Mononuclear Cells (PBMCs)
First, 15 cc of fresh heparin-containing venous blood was obtained from healthy volunteers. Then, under sterile conditions, an equal amount of phosphate-buffered saline (PBS) was added. Five mL of Ficoll (Inno-train, Germany) was added to 5 mL of blood mixed with PBS in a 15-cc tube. Blood was slowly added, eventually forming two separate phases. It was then centrifuged at 2000 rpm for 30 minutes. After that, a white, cloudy halo formed in the middle, which are PBMC cells. Platelets and plasma were elevated, and red blood cells and granulocytes were preci-pitated. Finally, PBMCs were gently removed with a Pasteur pipette, and cell survival percentage was counted after staining with trypan blue. PBMCs were then cultured in a flask containing RPMI 1640 (Roswell Park Memorial Institute) (Gibco, USA), FBS 10% (Fetal Bovine Serum) (Gibco, USA), and Pen-Strep 1%. The flask was incubated at 37°C and 5% CO2. Monocytic populations were separated from floating cells in a complete cell culture medium by the adhesion technique. Mononuclear cells were attached to the culture medium and transformed into macrophage-like monocyte cells, which confirmed the morphology of macrophage-like monocytes using a contrast phase microscope. After 3 days, the culture medium was changed, and the cells were passaged in three flasks separately, and each flask was treated (18).
It should be noted that the concentration of stimuli was previously determined by a cytotoxicity test using trypan blue. The results showed the use of Salmonella typhi concentrations for treatment in the authorized culture medium at a concentration of 4× 103 cfu/mL. Under completely sterile conditions, macrophage-like monocytes attached to the culture medium were separated from the flask by a scraper. After washing with serum-free medium (2000 rpm for five minutes), cell sediment was used to study gene expression in the next step.
RNA Extraction with TRIzol
TRIzol ™ Reagent Cat Numbers 155960 (Thermo Fisher Scientific, Inc.) was used to extract RNA from the flask. In addition, RNase / DNase free microtubes, filtered tips, and nuclease-free water were used to prevent DNA contamination. In addition, RNase enzyme (Yekta Tajhiz, Iran) was used to remove extracted RNA.
cDNA Syntheses
Smobio (Taiwan) was used for cDNA synthesis according to the kit instructions. 500 ng of RNA was removed from each sample and reached a volume of 4 μL with water. Then 0.5 μL of dNTPs and 0.5 μL of Oligo dt and Random hexamer primers were added and incubated at 70°C for 5 minutes in a thermocycler device. Then 2 μL of DTT, 2 μL of DEPC water, 0.5 μL of RNAse inhibitor and finally 0.5 μL of RT enzyme were added to each microtube. All microtubes were incubated in a thermocycler device for 50 minutes at 45°C.
Expression of TNF-α, IFN-γ using Real-Time PCR
For real-time PCR, Amplicon master mix and SYBR Green were used, which contains fluorescence and, after connecting to double-stranded DNA, emits fluorescent light. Finally, this light is measured by a Real-Time PCR device. In these reactions, the prepared cDNA was used as the template. GAPDH primer was used as an internal control. The sequence of primers used is shown in Table 1.
Statistical Analysis
All data obtained from Real-Time PCR were analyzed using Excel and GraphPad Prism software. Finally, the results were analyzed using the one-way ANOVA test, and statistical results less than 0.05 were considered significant.
Table 1. The sequence of primers for Real-time PCR
| Molecular weight | Sequence | Primer |
| 151bp | TTGGGTTCTCTTGGCTGTTA | IFN-γ F |
| TTCTGTCACTCTCCTCCTCCA | IFN-γ R | |
| 251bp | TTGGGTTCTCTTGGCTGTTA | TNF-α F |
| TTCTGTCACTCTCCTCCTCCA | TNF-α R | |
| 86 bp | TGCTGTCTCCCTGTTTGATGTATCT | GAPDH-F |
| TCTCTGCTCCCCACCTCTAAGT | GAPDH-R |
Sample Collection Results
Sixty patients with clinical signs of gastroenteritis, including 39 men and 21 women with a mean age of 55±4, were studied. Salmonella spp. was isolated from these 60 collected specimens, 3 of which were Salmonella typhi.
Table 2. Results of biochemical tests of fecal samples
| Trehalose | Rhamnose | Ornitin decarboxylase | Arabinose | Urease | LDC | citrate | VP | MR | Gas | H2S | kIA | Organism |
| + | - | - | - | - | + | - | - | + | - | + | Genus: Salmonella .Typhi |
Results of Specificity and Sensitivity of Primers
In order to design the primers, the mRNA sequence was extracted from the NCBI databases; the desired regions were designed for the target genes by Gene runner. Finally, the regions were specifically approved in the BLAST database.
Investigation of the Specific Amplification of Real-time PCR Products
The final products obtained from Real-time PCR were loaded on 5% agarose gel and electrophoresed to ensure the amplification of specific fragments of each gene and the absence of non-specific products and dimer primers. In addition, the melting curve was used (Figure 2).
Figure 2. Melting curve of studied genes such as TNF-α and IFN-γ, which indicates the specificity of their amplification.
Results of TNF-α Gene Expression
Quantitative expression of TNF-α gene in 60 cell culture media grown in medium containing Salmonella typhi or ATCC14028 standard strain with a concentration of 4×103 cfu/mL was measured by Real-Time PCR after RNA extraction and cDNA synthesis. The results showed that the relative changes in TNF-α gene expression in PBMCs treated with pathogen and ATCC strains both had a significant increase compared to the control sample (p: 0198). Also, the one-way ANOVA test and Tukey's multiple comparison tests were used for the group’s comparison (Table 3). The increase in TNF-α gene expression in the pathogen group was statistically significant than in the control group.
Table 3. Comparison of TNF-α gene expression in target groups
| Tukey's multiple comparisons test | Mean Diff. | 95% CI of diff. | Significant? |
| Control vs. ATCC | -0.4589 | -1.948 to 1.030 | No |
| Control vs. Pathogen | -1.875 | -3.364 to -0.3861 | Yes |
| ATCC vs. Pathogen | -1.416 | -2.905 to 0.07280 | No |
Quantitative results of changes in TNF-α gene expression in control samples and in PBMCs treated with the pathogen and ATCC14028 strains are shown in Figure 3, and the multiplication of this gene (folds) in each sample is shown in Table 4.
Figure 3. Changes in TNF-α gene expression in control, ATCC, and pathogen samples
Table 4. Expression TNF-α gene
| Sample Type | Fold Change |
| control | 1 |
| ATTC | 1.45 |
| Pathogen | 2.87 |
Results of IFN-γ Gene Expression
Quantitative expression of IFN-γ gene in the control group, samples treated with the pathogen and ATCC1609 strains was measured by Real-time PCR after RNA extraction and cDNA synthesis. The results showed that the relative changes in IFN-γ gene expression in PBMCs treated with the pathogen and ATCC14028 strains both had a significant increase compared to the control sample (P=0.001) (Figure 4). Also, using the One-way ANOVA test and Tukey's multiple comparison test, the groups were compared with each other, and the results are shown in Table 5. A statistically significant increase in IFN-γ gene expression in the pathogen group was observed than the control and ATCC 14028 groups.
Table 5. Comparison of IFN-γ gene expression in control, ATCC 14028 and pathogen
| Tukey's multiple comparisons test | Mean Diff. | 95% CI of diff. | Significant? |
| Control vs. ATCC 14028 | -0.1777 | -1.103 to 0.7475 | No |
| Control vs. pathogen | -2.010 | -2.935 to -1.084 | Yes |
| ATCC vs. pathogen | -1.832 | -2.757 to -0.9067 | Yes |
Quantitative results of changes in IFN-γ gene expression in control samples and in PBMCs treated with the pathogen and ATCC 14028 strains are shown in Figure 4, and the multiplication of this gene (folds) in each sample is shown in Table 6.
Table 6. Multiplication table of INF-γ gene expression
| Sample Type | Fold Change |
| Control | 1 |
| ATCC 14028 | 1.17 |
| Pathogen | 3 |

Figure 4. Changes in IFN-γ gene expression in control, ATCC, and pathogen groups
Studies have shown that disease progression is associated with changes in expression at the genome level. Although the molecular mechanism involved due to these genetic changes is not yet fully under-stood, researchers have reported the activation of several genes in different cellular pathways. The range of changes in the expression of these genes and the pathways involved in bacterial infection is very diverse and wide. In other words, disease development is a very complex process associated with the abnormal expression of several genes. Therefore, changes in gene expression can be used as a biomarker in the diagnosis of disease (19, 20). Therefore, these changes can be used to diagnose Salmonellosis and to study the mechanism of action of the immune system and bacteria against each other.
S. typhi pathogenesis, like other Salmonella, is complex and multifactorial. This microorganism cau-ses a wide and diverse set of infections (21). S. typhi pathogenicity depends on its ability to attack cells and form a protective lipopolysaccharide (LPS), the prese-nce of Vi antigen, and the production of invasin (inv genes), a protein that attacks non-phagocytic cells; where bacteria can survive and multiply intracellularly (22, 23). These factors in the host cause immune system reactions.
In the 1990s, IL-4 was discovered to have different effects on macrophage gene expression compared to IFN-γ and LPS. In contrast to classical macrophage activation by IFN-γ, IL-4-induced macrophage gene expression was described as "alternative activation". A few years later, in 2000, Mills et al. proposed a new classification of macrophages as M1 or M2. These terms derive from the observation of macrophage arginine metabolism in mice by working on T helper type 1 (Th1) and T helper type 2 (Th2) (24, 25).
Th1 that produces more IFN-γ showed macrophage activation in which nitric oxide (NO) is produced from arginine, whereas ornithine is produced from Th2, which produces IL-4 and TGFβ1. This finding led to a consensus in the scientific community that M1 macrophages (classical activators) exhibit inflamma-tory functions, while M2 macrophages (alternative activators) exhibit anti-inflammatory functions (26). In 2004, Mantowani et al. further subdivided M2 macrophages into subgroups M2a, M2b, M2c, and M2d based on stimuli applied and the resulting transcriptional changes. Classification of M1 / M2 macrophages is currently considered a fairly simple method that does not adequately describe the population spectrum of macrophages (27).
For example, identifying tumor-associated macro-phages (TAMs), which do not fit well into the M1 or M2 macrophage criteria, complicates the system. In addition, macrophages expressing T cell and CD169 receptors have been identified. The phenotype of macrophage subsets M1 and M2 and the function of macrophages derived from monocyte precursors are differentiated depending on the local tissue environ-ment (28). They respond to environmental signals in tissues such as damaged cells, activated lymphocytes, or microbial products to distinguish distinct functional phenotypes. The M1 macrophage phenotype is characterized by the production of high levels of proinflammatory cytokines, the ability to resist pathogens, strong antimicrobial properties, the production of highly reactive nitrogen and oxygen mediators, and the enhancement of Th1 responses. In contrast, M2 macrophages are characterized by their efficient control of parasitic infection, tissue regener-ation, immune regulation, tumor promotion, and efficient phagocytic activity (25). Therefore, in this study, the expression of genes related to cytokines TNF-α and IFN-γ was prioritized.
LPS, IFN-γ, and granulocyte-macrophage colony-stimulating factor (GM-CSF) polarize macrophages toward the M1 phenotype, which secretes large amounts of cytokines such as IL-1-β, TNF, IL-12, IL- 18, and IL-23. This helps to properly carry out the inflammatory responses of Th1 and Th17 cells specific for antigens (29). Typically, M1 macrophages stimu-late high levels of class II tissue adaptation complexes, CD68 markers, and CD80 and CD86 molecules. M1 macrophages have also been shown to regulate SOCS3 expression and activate NOS2 to produce NO from L-arginine. In the context of disease, M1 macrophages are involved in initiating and maintaining inflamm-ation, so they can be harmful to health (30).
Macrophages M1 and M2 have different chemokine and chemokine receptor profiles, with M1 secreting Th1-absorbing chemokines such as C1 in CXCL9 and CXCL10, and M2 secreting CCL17, CCL22, and CCL24. Recently, it has been shown that macrophages are able to repolarize from M2 to M1 in vitro, and reverse polarization depends on the chemokine environment.
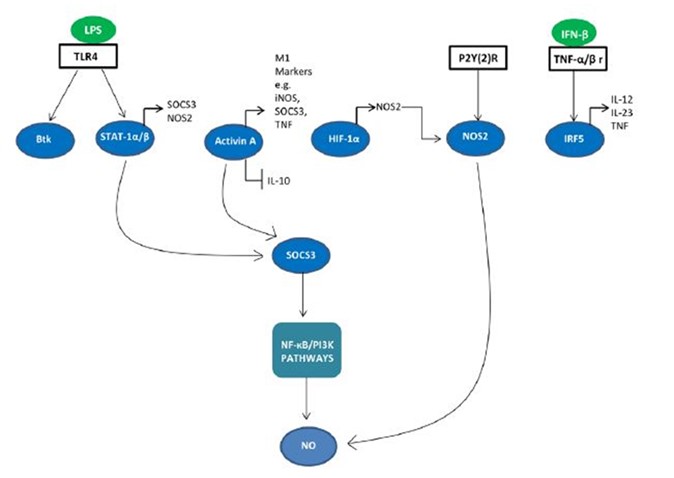
Signaling molecules are involved in M1 / M2 polarization. A network of transcription factors and post-transcriptional regulators are involved in M1 / M2 polarization (Figure 5). Interferon regulatory factor (IRF), STAT, and SOCS are all involved in the deviation of macrophage function towards the M1 or M2 phenotype (31, 32). IRF / STAT pathways activated by IFNs and TLR signaling polarize macrophages via STAT1 to M1 activation mode (33). M1 macrophages have been shown to regulate IRF5, which is involved in M1 polarization for M1 polarization and the whole STAT1-alpha / beta in an independent manner via MyD88 (27).
Figure 5. Signaling molecules involved in M1 polarization
In the present study, the gene expression of IFN-γ and TNF-α cytokines was evaluated to confirm the polarization of Salmonella typhi-stimulated macro-phages in vitro in two groups of clinical and standard strains of S. typhi. The results of gene expression analysis showed both genes have increased in expression in clinical and standard strains of S. typhi compared to the control group. This increase in expression was significant in the clinical strain of S. typhi, which was higher than the standard strain.
In a study conducted by Ami Febriza in 2020 to measure TNF-α on blood samples of S. typhi-infected mice using the ELISA method, it was found that TNF-α levels in infected samples increased and treatment of mice with antibiotics leads to its reduction (34). In addition, IFN-γ has been shown to play a vital role in resistance to S. typhi infection by increasing the antibacterial activity of macrophages. It has also recently been shown that IFN-γ and TNF-α activate the JAK / STAT1 / IRF pathway in mice infected with COVID-19 infection, leading to lethal cytokine shock, and therefore death from COVID-19 can be prevented by using neutralizing antibodies against them (14). In a study conducted in 2019, the relationship between the profiles of IFN-γ and TNF-α-producing cells and liver and kidney damage in hepatitis B virus infection was found (35). As a result, according to the studies, changes in the expression of IFN-γ and TNF-α cyto-kines can be used to identify S. typhi-related infections (36).
However, it should be noted that the present study could not investigate all the molecular aspects of polarization of macrophages and cytokines in the study and it is suggested that for better understanding of molecular pathways, more experiments be perfor-med.
The increased expression of IFN-γ and TNF-α in both clinical and standard strains of S. typhi compared to the control group confirms their role in macrophage polarization. Therefore, we can use their expression changes as a molecular biomarker in the diagnosis of Salmonella-related infections.
The authors would like to thank all the people who helped in the various stages of this research. The Department of Microbiology and Immunology, Faculty of Veterinary Medicine, University of Tehran is also commended for providing the location and experi-mental materials, as well as the Department of Laboratory Sciences of the Islamic Azad University, Boroujerd Branch.
This article is taken from the PhD thesis in bacteriology, which was done in collaboration with the University of Tehran Research Council, and the relevant resources were provided by the Vice-Chancellor for Research, University of Tehran.
Conflicts of Interest
There is no conflict of interest among the authors.
Received: 2021/07/28 | Accepted: 2021/11/5 | ePublished: 2021/12/8
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |