BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1350-en.html
2- PhD in Microbiology, Department of Biology, Faculty of Biological Sciences, Islamic Azad University, North Tehran Branch, Tehran, Iran
3- Assistant Professor of Genetics, Department of Genetics, Faculty of Basic Sciences, Islamic Azad University, Tehran, Iran , mitra_salehi_microbiology@yahoo.com
Staphylococcus aureus is one of the most important known causes of serious and deadly infections in hospitals (1). Nowadays, S. aureus is recognized as an important pathogen (2). S. aureus causes a wide range of diseases. Due to genome mutation potential, it has undergone many genetic changes. Because it has a flexible genome, pathogenic and resistant strains have expanded. Colonization of S. aureus, especially meth-icillin-resistant strains, is the cause of S. aureus prevalence and mortality (3-5).
Bacterial resistance to antibiotics is one of the challenges threatening human health. In recent deca-des, the use of antibiotics and medicine in agriculture has increased significantly and has also affected the environment (6,7). Drug resistance to antibiotics can be considered a major global threat posed by bacterial resistance following the development of resistance plasmid genes in bacteria. (8,9).
After the discovery of penicillin, it was initially used to treat staphylococcal infections. Still, the resistance of bacterial strains is increased so that in 1950, only 40% of hospital strains became resistant to penicillin, and this rate per the year 1960 reached 80% (10, 11). The cause of this phenomenon was the production of penicillinase by a bacterium that breaks down peni-cillin; Therefore, current antibiotics, such as penicillin-nase-resistant penicillin (such as oxacillin and methi-cillin), are used. Unfortunately, bacteria have become resistant to these antibiotics (12, 13).
Resistance to methicillin and other penicillinase-resistant penicillin is due to the presence of the mec operon. This operon is part of the staphylococcal chromosome cassette (SCCmec). The mecA gene encodes a penicillin-binding protein called PBP2a, which has a low affinity for beta-lactam antibiotics (7).PBP proteins are involved in the construction of bacterial cell walls (12). Therefore, the presence of such a new protein will not be affected by antibiotics, and the bacteria will easily survive. These strains are called methicillin-resistant S. aureus (MRSA). Another antibiotic called vancomycin and a new antibiotic called linezolid has been used for treatment (14).
The spa is one of the surface proteins of S. aureus. In addition to being a virulence factor of the bact-erium, it is used to determine the specific identity of S. aureus. With the molecular typing of this protein, it is possible to prevent epidemics, reduce the number of infections, and reduce the cost of nosocomial infections (15).
The presence of similar patterns in the spa gene indicates a common source of infection in hospital cases. Analysis of these patterns can help break the infection transmission chain in the hospital (16). Variation in S. aureus strains can improve the respo-nse to treatment. Identifying strains can be effective in how antibiotics are selected, so molecular typing is important to determine their characteristics and differentiate (17).
In this study, after extracting the genome of S. aureus from burn patients, (Staphylococcal protein A) spa and Panton-Valentine Leukocidin (pvl) genes were identified by PCR. The method of sensitivity and resis-tance to antibiotics in these strains was determined using the Disk-diffusion test. Then, the possibility of a relationship between pathogenic genes and antibi-otics was investigated.
Collection and Storage of Staphylococcus aureus
In this study, 56 samples of S. aureus positive from burn patients were collected from Shahid Motahari Hospital in Tehran, Panje- Azar Hospital in Gorgan, and Shahid Zare Hospital in Sari in 2018. These samples were transferred to blood agar and Mannitol salt agar medium and incubated at 37°C for 24 hours. Bioche-mical tests including gram staining, coagulase, catalase, Danse, and Mannitol fermentation were used to identify bacterial species.
Antibiogram Test
Antibiogram test was performed by Disk Diffusion test according to CLSI instructions. For this purpose, several bacterial colonies were removed and dissolved in physiological saline to equal the standard turbidity of half McFarland. Then agar was cultured on Müller Hinton medium, and antibiotic disks were placed on the culture medium at a standard distance and incubated at 37°C, and after 24 hours, the results were read.
Antibiotic discs of 30 μg vancomycin, 10 μg gentamicin, 5 μg methicillin, 1 μg oxacillin, 30 μg cefoxytin, and 15 μg erythromycin were prepared from Padtan Teb. S. aureus standard ATCC25923 was used as a positive control of the experiments.
DNA Extraction and PCR
Cinna Pure DNA bacteria kit (Cinna Pure DNA KIT-PR881614) was used for DNA extraction. SCCmec typing was performed for 56 MRSA strains using Multiplex PCR. For this purpose, the primers shown in Table 1 were used. Each PCR was performed in a final volume of 15 μL consisting of 2 μL primer, 1.5 μL of PCR buffer (10X), 75 μL of MgCl2, 0.6 μL of dNTPs, and 0.3 μM of NF Water. For each of the nine primers, 0.25 μL of DNA Ex-Taq polymerase and 7.3 μL of distilled water were used. For PCR, denaturation was perfor-med at 94°C in 15 minutes, followed by ten denaturation cycles, and for reconnection for 45 seconds at 94°C.
Table 1. Primer sequences used for Multiplex PCR testing for typing and determination of MSRA SCCmec subunit
| Primers | Oligonucleotide sequences (50–30) | Concentration (mM) | Amplicon size (bp) | Specifity |
| Type I-F Type I-R |
5´-GCTTTAAAGAGTGTCGTTACAGG-3´ 3´-GTTCTCTCATAGTATGACGTCC-5´ |
0.2 | 613 | sccmec I |
| Type II-F Type II-R |
5´-CGTTGAAGATGATGAAGCG-3´ 3´-CGAAATCAATGGTTAATGGACC-5´ |
0.2 | 398 | SCCmec |
| Type III-F Type III-R |
5´-CCATATTGTGTACGATGCG-3´ 3´-CCTTAGTTGTCGTAACAGATCG-5 |
0.2 | 280 | SCCmec |
| Type IVa-F Type IVa-d |
5´-GCCTTATTCGAAGAAACCG-3´ 3´-CTACTCTTCTGAAAAGCGTCG-5´ |
0.2 | 776 | SCCmec I |
| Type IVb-F Type IVb-R |
5´-TCTGGAATTACTTCAGCTGC-3´ 3´- AAACAATATTGCTCTCCCTC-5´ |
0.2 | 493 | SCCmec |
| Type IVc-F Type IVc-R |
5´-ACAATATTTGTATTATCGGAGAGC-3´ 3´-TTGGTATGAGGTATTGCTGG-5´ |
0.2 | 200 | sccmec |
| Type IVd-F Type IVd-R |
5´-CTCAAAATACGGACCCCAATACA-3´ 3´-TGCTCCAGTAATTGCTAAAG-5´ |
0.2 | 881 | SCCmec |
| Type V-F Type V-R |
5´-GAACATTGTTACTTAAATGAGCG-3´ 3´-TGAAAGTTGTACCCTTGACACC-5´ |
0.2 | 335 | SCCme |
| MecA147-F MecA147-R |
5´-GTGAAGATATACCAAGTGATT-3´ 3´-ATGCGCTATAGATTGAAAGGAT-5´ |
0.2 | 147 | mecA |
Isolation and detection of strains from clinical specimens
In this study, a total of 56 samples of sputum, blood, wounds, urine were collected from burn patients in Tehran, Gorgan, and Sari. The frequency of samples by type of sample is listed in Table 2. Using Grams staining, mannitol medium, Salt agar, catalase, and coagulase enzyme, 56 collected samples were found to be infected with S. aureus.
Table 2. Frequency of clinical samples based on sample type
| Urinary | Ulcer | Blood | Phlegm | Origin of sampling |
| 15 | 31 | 4 | 6 | N=56 |
| 26.7 | 55.3 | 7.1 | 10 | Percent |
Assessment of Microbial Susceptibility
Antibiogram results showed that 37 of the 56 strains studied were resistant to the antibiotic cefoxitin. In this study, evaluation of the pattern of antibiotic resistance of S. aureus strains showed that there was the highest resistance to methicillin antibiotics (100%) followed by cefoxitin (60%) and the lowest resistance to vancomycin (15%). Also, 47% of strains were resistant to oxacillin (Table 3).
Spa Gene Amplification Results
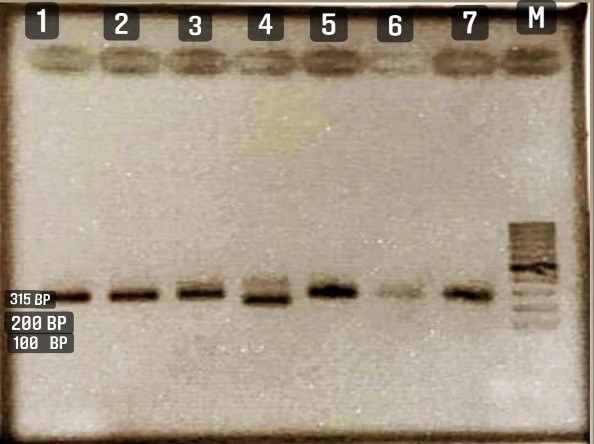
Figure 1 demonstrates the PCR results for the spa gene in S. aureus samples isolated from the patient. The results showed that the spa distribution was present in 82% (46 samples). Out of 46 samples, 40 samples belonged to wound samples.
Table 3. Results of microbial susceptibility assessment
| Oxacillin | Methicillin | Vancomycin | Gentamicin | Erythromycin | Cefoxitin | Antibiotics |
| 24 | 0 | 49 | 34 | 27 | 19 | Number of sensitives |
| 32 | 56 | 7 | 22 | 29 | 37 | Number of Resistances |
Figure 1. Electrophoresis results of PCR product for spa gene of lane M; marker bp100, lane1; standard strain, ATCC25923. Lanes 2 to 7 show clinical S. aureus strains with 315bp fragment length.
Results for pvl Gene Amplification
In order to investigate the presence of the PVL gene in S. aureus strains, specific primers of this gene were used, and the results were evaluated by a 180 bp band (Figure 2). The results showed that the pvl gene is observed in 12% of samples (7 samples).
In this study, no significant relationship was found between the presence of the pvl gene and sample type and pathogenic spa gene.
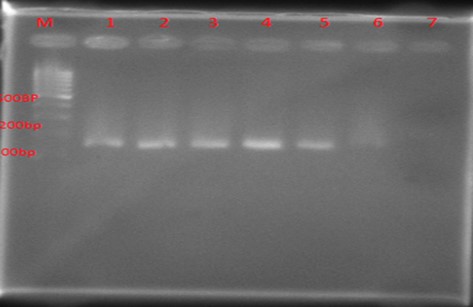
Figure 2. Results of PCR product electrophoresis on gel electrophoresis for pvl gene: 100 bp marker was used in lane M. Lane1 was a standard strain, 29213 ATCC. Lanes 2 to 6 strains of S. aureus clinically containing pvl gene (fragment length 180).
SCC mec Typing Results
Of the 56 strains analyzed by PCR, 100% were positive for the mecA gene. Type SCCmec in S. aureus were examined to determine the type of SCCmec MRSA isolates. As the findings showed, 16 (28.5%), 10 (17.8%), 5 (8.9%), 7 (12.5%) and 2 (3.5%), 3 (5.3%), 0 (0%), 3 (5.3%) MRSA isolates contained SCCmec type III, type I, type II, type Iva, type IVb, type IVc, type IVd and type V. In addition, 10 isolates (17.8%) could not be typed. Our study also showed that the number of HA-MRSA-related strains was higher than CA-MRSA in the isolates under study (Figure 3).
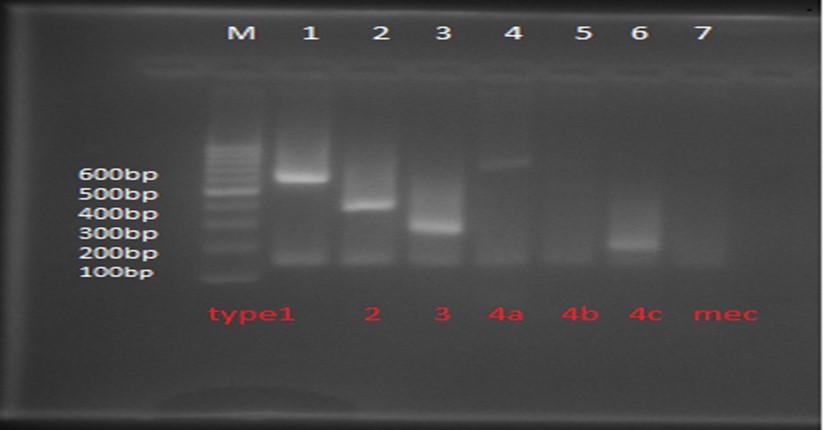
Figure 3. Multiplex PCR agarose electrophoresis. Lane 1, type SCC mecI (613 bp), Lane 2, SCC mec type II (398 bp), lane3, SCC mec Type III (280 bp), lane 4, SCCmec type Iva (776bp), lane 5, c Type IVb (493bp) SCC me, lane 6, type IVc (200bp) SCCmec.
S. aureus is one of the most important human pathogens. It has been a major cause of hospital infections over the past decades (18, 19). Prompt detection and treatment of MRSA infections are the most important measures to prevent the spread of infection and reduce the risk of mortality in patients (12). MRSA infections are divided into two groups: HA-MRSA and CA-MRSA. These classifications can be identified by the source of mobile genetic elements, SCCmec. (20,21).
SCCmec is the most important determinant of the source of MRSA infection. If HA-MRSAS or CA-MRSA is detected, we will manage MRSA infections and choose the best treatment protocol (22). Each type of SCCmec has unique genetic elements, and the determination of the mecA gene by PCR is more sensitive and accurate than the oxacillin disk test (23). Therefore, the mecA gene was identified by PCR. According to the results, the presence of the mecA gene in 56 studied strains was 100%. At the same time, oxacillin resis-tance was more than 50% (24), which is comparable to the results of the study of Ionescu et al. Ionescu research showed in 57 strains of S. aureus, phenotypically 22 strains were resistant to oxacillin, and 28 strains contained mecA gene (25). In another study by Hammad et al., All MRSA strains isolated from clinical specimens contained the mecA gene (26).
Also, antibiotic susceptibility testing showed that 12.5% of the samples were sensitive to vancomycin. Resistance to oxacillin was less than 60%, and gentamicin, amikacin, cephalexin, cefoxitin, clindamy-cin, and erythromycin was between 30% and 70%. However, some antibiotics, such as vancomycin, are still effective for treatment.
In this study, the highest resistance was to oxacillin, methicillin and erythromycin. Also, five types of SCCmec were identified, and among them, type III was the most common in isolates, and multi-drug resistance was higher in type I and III strains than in other types. A similar study in 2016 in northern Iran showed that only SCCmec type III had the highest drug resistance (13). In this study, the other type was SCCmec I (17.8%) and type IVa (12.5%) (27).
A 2009 study by Vazquez on a variety of samples in Spain found that the highest percentage of isolates belonged to SCCmec type IV, while in a similar study in 2019 by Zhanget al, performed clinical trials and the dominant type and SCCmec type III were reported (28). Some researchers examined SCCmec typing patterns in different samples, except for the 2011 study by Goss and Muhlebach in which patients with cystic fibrosis were selected as the study population, most had SCCmec type I. According to other studies, SCCmec types I, II, III are known as HA-MRSA, while CA MRSA strains are associated with SCCmec types IV and V (16). The prevalence of HA-MRSA in this study was 57.7 (55 patients), higher than CA-MRSA. In this regard Rainard et al, and Abdi et al. (17, 18) have reported similar cases.
This study aimed to obtain the epidemiology and SCCmec typing of MRSA in patients in three different hospitals in Tehran, Gorgan, and Sari. Of the 56 MRSA isolates studied during 2019 using Multiplex PCR, SCCmec type III (28.5%) and SCCmec type I (17.8%) were studied. The present study results indicated that MRSA isolates may have originated from HA-MRSA or clonal aberration of MRSA.
The results of this study showed high resistance to methicillin among S. aureus isolates. Therefore, to select an appropriate antibiotic to treat S. aureus infections and prevent antibiotic resistance, doctors should prescribe appropriate antibiotics based on their effectiveness and availability.
The pvl gene causes permeability and antibiotic resistance in this bacterium. The spa gene also causes adhesion and increases bacterial resistance, and has a high polymorphism whose unique number and sequence of replication varies among strains. Mole-cular typing of this protein can investigate the possi-bility of a common source of infection that circulates or is transmitted from person to person in hospitals.
None.
None.
Conflicts of Interest
There is no conflict of interest between the authors.
Received: 2021/05/30 | Accepted: 2021/09/1 | ePublished: 2021/12/8
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |