BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1271-en.html
.
Salmonellosis is one of the most common food-borne infectious diseases in the world, particularly in developing countries. It can be said that this disease imposes many damages to the countries annually due to the subsequent health problems (1). This disease is caused by various serotypes of Salmonella bacteria that are highly diverse, biochemically and serologically (2). Salmonella infantis has been reported as the most common isolated serotype in recent years (3, 4). Vegetables, dairy products, infected poultry, meat and eggs are among the most important sources of infection in humans (5, 6, 7). According to the World Health Organization (WHO), 17 million people are annually diagnosed with typhoidal Salmonella. Nearly 600,000 of them die, and 1.3 billion acute gastroenteritis or diarrhea cases are caused by non-typhoidal Salmonella leading to three million deaths and making salmonellosis a major health issue, globally (8).
Antibiotic therapy is one of the important methods applied to reduce and control Salmonellosis in birds and humans (9, 10). Antibiotic resistance to this bacterium has been increased due to the excessive consumption of antibiotics in the fields and health centers, so that the spread of resistant strains has become a global problem (11, 12), and a serious threat to public health (13, 14).
Considering that poultry products, especially chicken, count for sources of Salmonella infection, the present research was carried out about contamination of chicken to Salmonella serotypes in Ardabil region. Therefore, the aim of this study was to evaluate Salmonella infection in chicken to identify the regarding serotypes and to determine drug resistance to 10 antibiotics which are commonly used in Iran.
Sample Collection
This cross-sectional descriptive study was carried out within five months in 2017 in cooperation with the Veterinary Organization of Ardebil province. In this study, chicken supply and distribution centers with health license in Ardabil city were included. The study consisted of 100 chicken samples that were randomly selected from above mentioned centers from four districts of north, south, east and west of the city. Samples were collected in sterilized 30-gram plastic containers and transferred to the laboratory.
Salmonella Identification
Samples prepared in selenite F rich medium (Himedia, India) were cultured at 37°C for 24 hours and transferred and cultured in the selected solid mediums of Salmonella such as MacConkey Agar (Merk, Germany), Salmonella-Shigella Agar (Himedia, India) and Brilliant Green Agar (Himedia, India). The colonies were investigated after incubation for 24 hours at 37°C. Negative lactose colonies (uncolored) were considered as suspected colonies with and without H2S production. Suspected samples were again cultured in xylose lysine deoxycholate (XLD) agar (Himedia, India), which produced red and black centered colonies. Suspected colonies were cultured in differential mediums including Urea Broth, Simon Citrate Agar, SIM, TSI Agar (Quelab, Canada) and their reactions were investigated in sugar mediums including MRVP, mannitol, maltose, lactose, arabinose, sucrose, glucose and D-xylose (Quelab, Canada) (15). Then, by comparing the results with the biochemical table, the isolated bacteria were determined. Determination of serogroups and serotypes of positive samples was performed using serological tests and polyvalent anti-serums (Mast, England) (A to D). Numerous monovalent antiserums available for the diagnosis of serum group O were used. Subsequently, to determine the serotype within the group, flagellar antigens (H2, H6, HL, Hgm) were used according to the instructions of Mast Company (England), using the slide agglutination method (15). For this purpose, a concentrated suspension of bacteria was prepared in a physiological serum on a slide, then a drop of monovalent O serum was added to it and agglutination was evaluated in less than two minutes. Then, the above mentioned sample was determined with H anti-serums (phases 1 and 2) and by observing agglutination and based on the Kauffman-White table of bacteria serotype.
DNA Extraction
The boiling method was employed to extract the DNA. For this purpose, confirmed colonies, were directly cultured in a peptone water medium and after 24 hours, 100 μL of bacterial culture was boiled in the peptone water medium for 10 minutes with 500 μL of distilled water. After centrifugation of the samples in 14000 rpm, supernatant was used as DNA.
PCR Test
To determine the molecular characteristics of different Salmonella species, PCR was performed. Firstly, PCR was performed using St11/St15 primers to determine the genus of Salmonella, and then PCR test was also performed to determine Salmonella typhimurium (S. Tm) by using specific primers (fliC) (Table 1). S. Tm ATCC 14028 was used as positive control. The details of the sequences of their returning and forward primers are as follows:
Table 1: Primers Used for PCR detection of Salmonella and Salmonella Tm
| References | Amplicon Size (bp) | Primer sequence (5' to 3') | Target gene |
|---|---|---|---|
| Soumet etal. (1999) (16) | 429 | GAGCCAACCATTGCTAAATTGGCGC A | ST 11 |
| 429 | GGTAGA.AATTCCCAG.CGGGTACTGG | ST 15 | |
| Azizpour A (2020) (5) | 432 | CCCGCTTACAGGTGGACTAC | fliC |
| 432 | AGCGGGTTTTCGGTGGTTGT | fliC |
The PCR was performed in 50 µl volume containing 5 µl each Primers (Metabion, Germany), 1 µl of Taq Polymerase, 0.5 µl of dNTP, 1 µl of MgCl2, 5 µl of 10x Loading DNA, 5 µl of DNA template and 32/5 µl of sterile distilled water. The steps of the thermocycler device (Eppendrof, Germany) were as follows: Initial denaturation at 94°C for 5 minutes and 35 cycles for the other steps containing a denaturation at 94°C for 45 seconds, annealing at 56°C for 45 seconds, extension at 72°C for one minute and Final extension at 72°C for 5 minutes. Electrophoresis was performed for the amplified products (20 μl) on 1.5 % agarose gel (Simga, USA) prepared in 1x TBE buffer (89 mmol 1-1Tris, 89 mmol 1-1 Borate and 2 mmol 1-1EDTA) at 80 V for 45 minutes. This gel was stained with DNA safe SYBR (SinaClon, Iran) and then photographed under UV light. The images were prepared using a Gel Doc system (Cambridge, Germany).
Antibiotic susceptibility testing
After determining the serogroups of Salmonella isolates, their antibiotic resistance patterns were investigated using antibiotic disks manufactured by Padtan Teb Company of Tehran, Iran by Disc diffusion, Kirby Bauer methods according to instructions of Clinical and Laboratory Standards Institute (5). The tested antimicrobial agents and their concentrations were as follows: tetracycline (30 µg), trimethoprim-sulfadiazine (1.25/23.75 µg), florfenicol (30 µg), enrofloxacin (5 µg), co-trimoxazole (trimethoprim-sulfamethoxazole) (1.25/23.75 µg), amoxicillin (10 µg), ciprofloxacin (5 µg), ampicillin (10 µg), chloramphenicol (30 µg) and doxycycline (30 µg). To perform an antibiogram test, a standard 0.5 McFarland solution was prepared from the suspension and then cultured in Mueller-Hinton agar medium. In the next step, the disk locating was performed and the plates were incubated at 35°C for 18 hours (15). Finally, by measuring the diameter of zone of inhibition, the bacterial resistance or sensitivity to tested antibiotics was determined.
Of the100 chicken samples, S. enterica was isolated in six cases (6%), from which, four isolates (66.6%) were assigned to serogroup C, one isolate (16.6%) was assigned to serogroup D, and also one isolate (16.6%) to serogroup B. Of the four isolates of the serogroup C, three isolates were assigned to the S. infantis serotype and one isolate was assigned to the S. thompson serotype. One isolate of the serogroup D was assigned to S. enteritidis, and one isolate of serogroup B was considered as S. Tm.
The results of the study of the antibiotic sensitivity and resistance of Salmonella isolates to 10 common antibiotics are shown in Table 2. All isolates were sensitive to two antibiotics, enrofloxacin and ciprofloxacin. More than 60% of the isolates were resistant to 7 antibiotics. The only isolate assigned to the serogroup B was sensitive to doxycycline, florphenicol, enrofloxacin and ciprofloxacin and resistant to six antibiotics. The isolate assigned to the serogroup D was fully sensitive to florphenicol, enrofloxacin and ciprofloxacin and fully resistant to tetracycline, doxycycline, chloramphenicol, amikacin, sulfadiazine+trimethoprim, co-trimoxazole and ampicillin. The isolates assigned to the serogroup C were fully sensitive to enrofloxacin and ciprofloxacin. The resistance of these isolates was 75% against four antibiotics; doxycycline, chloramphenicol, sulfadiazine+trimethoprim and ampicillin, and 50% against tetracycline, amikacin and co-trimoxazole and 25% against florphenicol.
Table 2: Resistance and sensitivity of Salmonellas isolated from chicken meat indicated by serogroup
| Serum Groups Antibiotics |
B | D | C | C | C | C |
|---|---|---|---|---|---|---|
| Tetracycline | R | R | R | S | S | R |
| Doxycycline | S | R | R | R | S | R |
| Florphenicol | S | S | R | S | S | S |
| Chloramphenicol | R | R | R | R | R | S |
| Enrofloxacin | S | S | S | S | S | S |
| Ciprofloxacin | S | S | S | S | S | S |
| Amikacin | R | R | R | S | R | S |
| Sulfadiazine+Trimetoprime | R | R | R | R | R | S |
| Cotrimoxazole | R | R | R | S | R | S |
| Ampicillin | R | R | R | R | S | R |
R: Resistance S: Susceptible
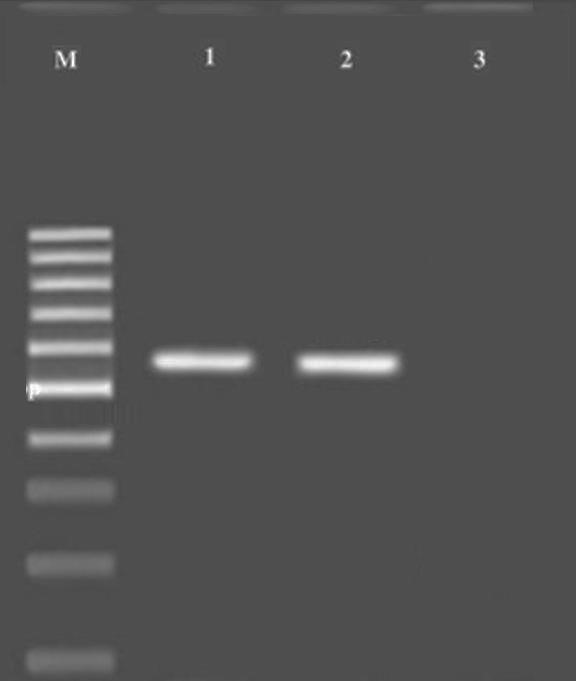
S. Tm PCR product was detected on 1.5% agarose gel, and the 432 bp band was observed. The results of the PCR products on the agarose gel and the bands formed with the markers are shown in Figure 1.
Figure 1. Gel electrophoresis of the PCR products. M: Molecular ladder; 1: Positive control; 2: S. Tm band at 432 bp; 3: Negative control
Salmonellosis is one of the most important bacterial diseases of the poultry industry, especially broiler chickens in the world and the industrial poultry and its products are the most important sources of transmission of Salmonella to humans (10, 17). Salmonella contamination occurs in humans as food poisoning, gastroenteritis, typhoid fever, and sometimes septicemia (6, 18). Therefore, identification of Salmonella, especially circulating serotypes and determinination of the drug resistance pattern in order to prevent the spread of resistance genes are of great importance in terms of public health.
In Ethiopia, Salmonella was isolated by 21.2% from 378 poultry carcasses in 2003 (12). In Nigeria, in 2005, 12.5% of 120 chicken feces samples were positive with Salmonella (13). In another study in 2007, out of a total of 336 carcasses in industrial slaughter-houses in northern Spain, 17.9% were infected with Salmonella (19). In Shanxi Province of China, 515 chicken samples were tested for Salmonella in 2010 and 54% contamination was announced (20). In Bangladesh, in 2011, Out of 503 poultry carcasses, Salmonella infection was reported in 37% of the cases (10). In the central region of Mato Grosso do Sul of Brazil in 2011, Salmonella was isolated by 11.28% from 258 slaughter poultry (21). In Algeria in 2012, out of 314 samples of studied red meat and chicken, 19.34% were positive with Salmonella (22). In a study in northeastern Poland, 300 slaughtered turkeys were investigated during 2014, of which, 8.3% were infected with Salmonella (1). In Egypt, Salmonella was also isolated by 34% from 200 investigated meat samples (23).
Various reports have been provided on Salmonella infection rates in different regions of the country. In Hamadan Province of Iran in 2000, 140 chicken samples were examined for Salmonella and the contamination rate of 8.6% was reported (24). A study conducted in Fars Province of Iran during 2005, resulted that 15.62% of 192 samples from broiler flocks were infected with Salmonella (7). In the southern regions of Tehran in 2007, out of 315 samples of chicken, 11.3% were infected with Salmonella (25). In 2009, 1125 poultry samples were collected from native poultry in northern Iran and the rate of infection with Salmonella was 2.4% (2). In 2010, Salmonella was isolated with the rate of 31% from 93 investigated chicken flocks in Ahwaz Region of Iran (26). In Uremia Region of Iran, in the year 2011, 20.8% Salmonella were isolated from 360 various organs of poultry (16). Out of 245 cloaca samples collected from slaughtered poultry in Arak Region of Iran, in the year 2013, 30.61% of the samples were infected with Salmonella (27). In Gilan Province of Iran, in 2013, Salmonella was isolated at a rate of 15% from 20 slaughtered broiler chicken flocks (3). In another study in the year 2013 in Talesh Region of Iran, Salmonella contamination in 100 chickens was reported to be 21% (28). In Chaharmahal and Bakhtiari Province or Iran, 620 samples of meat collected from supermarkets in 2014 were reported as contaminated with Salmonella by 4.51% (6). Another study in Ardabil Region of Iran, showed that 260 samples of chicken and viscera on the market were contaminated to Salmonella by 10% (15). In Alborz Province of Iran, in 2015, out of 560 chicken samples (liver, heart and gizzard) Salmonella was isolated at a rate of 19.22% (29). In another study, three poultry slaughter-houses in Alborz Markazi and Fars provinces of Iran were investigated during 2016 and this study showed that of the 585 samples, 25.5% were infected with Salmonella (30). In the present study, the infection rate of chicken to various Salmonella species was found to be 6%. This infection rate is lower than the majority of the previous reports (14, 15) and higher that some of them (2, 6). This difference seems to be due to geographical location, health condition of poultry centers, type and volume of samples, and methods applied to detect contamination.
There are different reports on the prevalence of various Salmonella serogroups and serotypes in different regions of the world. In a study by Mehrabian et al. (2001) in Tehran Region of Iran on beef, chicken and egg samples, the results showed that S. enteritidis is dominant to other Salmonella species (31). In Spain, the highest serotypes isolated from poultry samples were S. enteritidis, Salmonella enterica serovar Typhimurium (S. Tm), S. virchow, S. newport, and S. heidelberg (32). In Thailand, S. Tm, S. gallinarum, S. pulorom and S. cholerasuis were reported as the most common serotypes (32). In a study by Sultan Dallal et al. (2007) in Tehran Region of Iran, the dominant serotype was S. thompson in chicken specimens (25). Namaei et al. (2009) isolated two local serotypes of S. enteritidis and S. Tm from eggs in Birjand Region of Iran (34). According to Shapouri et al. (2009) in Zanjan region of Iran, the highest serotypes isolated from egg shell were S. enteritidis (23.3%), S. agona (10%), S. virchow (10%), S gallinarum (6.7%), S. infantis (3.3%) and S. heidelberg (3.3%) (29). Azizpour (2018) isolated four Salmonella serotypes in poultry flocks in Ardabil city of Iran, which consisted of S. enteritidis (59.1%), S. infantis (31.8%), S. thompson (4.5% ) and S. Tm (4.5%) (4).
In a study by Zahraei Salehi et al. (2005) in Fars Province of Iran, 70% of Salmonella was assigned to serogroup D1, 20% to serogroup C1, 6.6% to serogroup C2 and 3.3% to serogroup B (7). Pooladgar et al. (2010) isolated three serogroups B, C, D in Ahvaz Region of Iran (26). Akbarmehr (2010) in Sarab region of West Azerbaijan Province of Iran showed that 53.3%, 26.6%, 11.1% and 8.8% of Salmonellas were assigned to groups D1, B, C1 and C2, respectively (9). In a study by Ezzatpanah et al. (2013) in Arak Region of Iran, the serogroup D1 of Salmonella was reported as the dominant serogroup (27). Raeisi and Ghiyami (2015) indicated that three serogroups of Salmonella (C, B and D with 92.3%, 3.8% and 3.8% isolation rates, respectively) were isolated from chicken and viscera in Ardebil Region of Iran. Several studies have also been conducted on the prevalence of serogroups in different countries (15). Goncagul et al. (2005) in Turkey, isolated the groups A, B, C and D on the skin of wings of slaughter poultry and represented that serum D was the majority (11). According to the results of Mahmud et al. (2011) in Bangladesh, 43% of isolates were assigned to serogroup B and 57% to serogroup C (10). In the present study, three serogroups C, B and D were isolated; serogroup C had the highest rate of isolates with an isolation rate of 66.7% and among isolated serotypes, S. infantis was the dominant serotype with the rate of 50%. The findings of this study on the dominancy of serogroup C and S. infantis are consistent with the results of some of researchers (9). This issue seems to be due to the endemic of this type of Salmonella in recent years in Iran (7, 27). Of noteworthy points in the present study, is the assignment of a high percentage of group C isolates to S. infantis which was the second highest Salmonella infection rate. Comparison of isolation rates from the results of present study with the results of Raeisi and Ghiyami’s study (2015) on chicken in Ardebil (15), indicates that there is a change in the prevalence rate of serogroups, so that serogroups D and B have been increased from the rate of 3.8% to 16.4%. While the serogroup C has been decreased from the rate of 92.3% to 66.7%. In other words, the highest prevalence of Salmonella in this study is also related to S. infantis serotype. So that, in recent years, it has been shown that the prevalence of Salmonella serogroup C, especially S. infantis serotype is increasing in broiler poultry flocks, which is important in terms of public health (15). Various results for serogroups can be due to differences in geographic regions and also time and type of sampling. It seems that serotypes can replace each other in a particular region and period, due to the existed rotational situation between serotypes (4).
Several reports have been provided on the antibiotic resistance of Salmonella in different countries, which indicates the diversity and high levels of drug resistance in different regions. Molla et al. (2003) in Ethiopia, indicated that among 23 tested antibiotics, the highest levels of resistance were related to sulfamethoxazole (51.2%), amoxicillin and ampicillin (46.2%), tetracycline (2/41%), chloramphenicol (30%), florphenicol (27.5%), streptomycin (22.5%) and cotrimoxazole (21%), and there was no resistance against nitrofurans, quinolones, cephalosporins, kanamycin and neomycin (12). In Spain, Capita et al. (2007) observed that the highest levels of resistance were against sulfonamides, fluoroquinolones and tetracyclines (19). According to reports by Yang et al. (2010) in Chinese Shanxi Province, the highest percentages of resistance were related to sulfamethoxazole+trimethoprim (67%), tetracycline (56%), nalidixic acid (35%), ciprofloxacin (21%) and ceftriaxone (16%) (20). In Chinese Sichuan Province, Ruichao et al. (2013) reported the highest percentages of resistance to tetracycline (77%), sulfamethoxazole+trimotropil (43%), nalidixic acid (41%), streptomycin (41%) and ampicillin (25%), and the lowest percentages to gentamycin (15%), amoxicillin (14%), ciprofloxacin (12%), and florphenicol (10%) (14). Abdelghany et al. (2015) in Egypt observed the highest percentages of resistance to nalidixic acid (98.8%), sulfamethoxazole+trimethoprim (96.4%), oxytetracycline (95.2%) and ampicillin (91.06%) (23).
In Iran, studies conducted in different regions of the country indicate the emergence and increase of drug resistance in the Salmonella isolates. So that in Zahraei Salehi et al. (2005) studies, all isolates were sensitive to colistin, gentamicin, ciprofloxacin, enrofloxacin and cefalotin, and the highest drug resistances were observed to be against streptomycin, flumequin, nalidixic acid, tetracycline, neomycin and trimethoprim (7). Peighambari et al. (2011) reported the highest resistances to tetracycline (66.6%), furazolidone (52.8%), nalidixic acid (43.8%), linoecspectin (42.3%), flumequine (40.6%) and streptomycin (39.1%), while ciprofloxacin and imipenem had 100% sensitivity (18). According to reports by EzzatPanah et al. (2013), the highest drug resistance percentages were observed when using nitrofurantoin (92.6%), nalidixic acid (86.7%), clistein (64%), tetracycline (54%), furazolidone (49.3%), and amoxicillin (45.3%) (27). Asadpour et al. (2013) showed that all isolates were resistant to tetracycline, streptomycin, nalidixic acid, cefazolin, and sulfamethoxazole+trimethoprim (3). Raeisi and Ghiyami (2015) observed the highest resistance rates to streptomycin and nalidixic acid (100%), tetracycline (92.3%), neomycin and furazolidone (84.6%) and chloramphenicol (73.3%). Also, the lowest resistance rates were against amoxicillin and ampicillin (11.5%), ciprofloxacin (7.7%) and gentamicin (3.7%) (15). Sodagari et al. (2015) in Alborz Province introduced tetracycline as the most resistant antibiotic against Salmonella isolated from poultry (29).
In the present study, among the common antibiotics in the poultry industry, the highest rates of resistance were against sulfadiazine+trimethoprim, tetracycline, doxycycline and florphenicol, respectively. In the group of human antibiotics, the highest resistance rates were observed against ampicillin, chloramphenicol, co-trimoxazole and amikacin, respectively. The resistance rates of isolates in the present study were different from the results of some previous studies, which can be due to long-term and excessive consumption of antibiotics in different regions of the country and the genetic transfer of drug resistance between bacteria (15, 27). In the present research, the lowest resistances were observed against fluoroquinolone and florphenicol groups. Therefore, these drugs may be effective in the treatment of salmonellosis.
According to the results of this investigation, the predominant serotype is S. infantis. Poultry Salmonella isolates are resistant to the majority of antimicrobial agents under study, which is of great importance in term of public health. This situation can be due to continuous consumption of drugs in poultry farms. Therefore, in order to prevent the emergence of resistant strains of bacteria, the principled consumption of antibiotics seems to be necessary.
The author of this study would like to thanks Dr Ciamak Ghazaei from University of Mohaghegh Ardabili who helped with performing this research.
This research resulted from an independent research without receiving any financial support.
Authors declare that there is no conflict of the interest.
Received: 2021/01/23 | Accepted: 2021/02/20 | ePublished: 2021/04/9
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |