BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1158-en.html
2- Department of Microbiology, Faculty of Science, Islamic Azad University, Kazeroun, Iran , Majidbaseri@hotmail.com
3- Hospital Infection Research Center, Isfahan University of Medical Sciences, Isfahan, Iran
Moraxella catarrhalis is a non-motile and non-pigmented gram-negative diplococcus considered as a nonpathogenic bacteria and respiratory system normal flora up until 1995, however; recognized as a human pathogenic agent afterwards (1). These bacteria can cause upper and lower respiratory tract infections in adults (1-3). Subsequent to Streptococcus pneumoniae and Haemophilus influenzae, M. catarrhalis is a leading bacterial infection agent responsible for 15% to 20% of the cases (4).
Due to their broad innate and induced resistance to numerous antibiotic families, treating such respiratory infections is troublesome for medical practitioners (3). These bacteria are known for their exclusive innate antibiotic resistance and very likely acquired antibiotic resistance (5). Antibiotic resistance has led to several difficulties M. catarrhalis infection treatment.
Innate resistance in the bacteria can be on account of drug enzyme inhibition such as inhibition of beta-lactamase which transforms β-lactam using hydrolysis, bacteria cell membrane permeability alteration which leads to drug resistance such as purines, and expressing active secretion systems or efflux pumps (5). Due to their broad substrate variety, efflux pumps in this family have been divided into five grand groups including RND (Division Nodulation Resistance) multidrug efflux pumps (6).
AcrAB-OprM is an efflux system of RND family in M. catarrhalis. The pump consists of an inner membrane pump (AcrB), an outer membrane channel (OprM), and a periplasmic adaptor protein (AcrA) that facilitates outer and inner membrane antibiotic transmission in the bacteria. As AcrAB-OprM substrates, drugs can elevate acrAB-oprM expression which can lead to multidrug resistance (5).
Efflux pumps not only increase minimal inhibition concentration (MIC) of antibiotics, but also develop drug-resistant lineages using proton motive force (PMF) and decreasing drug concentration (7, 8). With increasing efflux pump expression, these bacteria resist to a wide range of drugs. Their synergism with other drug resistance mechanisms is of great importance and should be studied like other resistance mechanisms (8-10). Recognition and inhibition of efflux pumps seem promising reinforcements for antibacterial substances efficacy. In the recent years, numerous inhibitors including natural products, various antibiotics, and synthetic molecules have been tested on gram positive and gram-negative bacteria in order to solve the resistance problem (11). Considering the ever-increasing bacterial resistance, and the outstanding role efflux pumps play in it, discovering resistance adjusting factors or more specifically, efflux pump inhibitors (EPIs) is of great importance so that designing novel drugs necessitates recognizing and understanding resistance inducing systems in efflux pumps (8, 9, 11).
Increasing antibiotic resistance in the bacteria is a global infectious and respiratory diseases risk. According to the absence of phenotypic and genotypic studies on the role of efflux pumps in M. catarrhalis infection resistance in Iran, this study aims to assess phenotypic and genotypic prevalence of acrAB-oprM efflux pump genes as significant antibiotic resistance agents in M. catarrhalis and inhibit efflux pumps using phenylalanine-arginine β-naphthylamide (PaβN) as a clinical practice.
Sampling and Identification of Bacteria
In this cross-sectional descriptive study, sampling was done on patients aged 3-75 years, both male and female, with respiratory infections and pneumonia referred to Bagherul Uloom clinic and laboratory and Valiasr Hospital clinic in Kazerun from December 2015 to May 2016. Inclusion criteria were patients referred for suspected tuberculosis, samples with respiratory infections, adult smokers with chronic bronchitis and sinusitis and children with acute middle ear infection. The above identification was made by a specialist.
Finally, 137 samples of sputum and throat swab (oral and laryngeal pharynx), and purulent secretions of the middle ear taken under standard conditions, were transferred to the laboratory. Samples were cultured in the laboratory for 24 h on chocolate agar and blood agar (Merck-Germany) growth mediums. Standard microbiological and biochemical tests to differentiate bacteria, including gram staining, oxidase, catalase, DNase, the glucose fermentation test (glucose, sucrose, maltose, fructose, and lactose), and nitrate reduction were used to identify and isolate M. catarrhalis bacteria (12).
Determination of Antibiotic Susceptibility by Kirby-Bauer Method
The pattern of antibiotic susceptibility in the strains obtained using the disk diffusion method (Kirby-Bauer) was investigated in accordance with the guidelines of the CLSI (Clinical and Laboratory Standards Institute) version 2016.
Turbidity equivalent to 0.5 McFarland tube was prepared and then cultured on the Mueller-Hinton-Agar growth medium (Merck, Germany) for 24 h and then antibiotic disks (Padtan Teb, Iran) were placed on the growth medium. Antibiotic resistance to penicillin (10 µg), ampicillin (10 µg), erythromycin (15 µg), chloramphenicol (30 µg), clarithromycin (15 micrograms), cefazolin (30 µg), coamoxyclav (10+20 µg), trimethoprim-sulfamethoxazole (1.25-23.75 µg), tetracycline (30 µg), ciprofloxacin (15 µg), azithromycin (15 µg), amikacin (30 µg), gentamicin (10 µg) and ceftazidime (30 µg) (Himedia, India) was investigated. After 24 h at 37°C, the diameter of the growth inhibition zone around the discs were measured and the strains were divided into susceptible, semi-susceptible, and resistant groups according to the CLSI guidelines. Isolates that showed resistance to more than two classes of antibiotics were introduced as strains with multiple MDR resistance (13).
Identification of Moraxella catarrhalis by PCR
Identifying M. catarrhalis with multidrug resistance was confirmed by the SrRNA16 sequence (Table 1). DNA of these samples was extracted by Kit (Yekta Tajhiz, Iran). PCR reaction was performed on a final volume of 25 μL including 18 μL of Mastermix (Yekta Tajhiz, Iran), 3 μL of deionized distilled water, 1 μL of each forward and reverse primer, and 2 μL of template DNA. PCR program included initial denaturation at 94°C for 5 min, 35 thermal cycles including degreasing at 94°C for 20 sec, primer binding at 60°C for 30 sec, amplification at 45°C for 72 sec and the final amplification at 72°C for 2 min.
Amplified DNA was sent to Macrogen Korea Company for sequencing, and the identified sequences were blasted for gene analysis.
Phenotypic Study of Efflux Pump Using Ethidium Bromide-Agar Cartwheel Method
In order to evaluate the phenotypic activity of efflux pumps, the isolates of M. catarrhalis were examined by Ethidium Bromide-agar Cartwheel method. First, ethidium bromide solution (Merck, Germany) was prepared in distilled water at a concentration of 50 mg/mL and stored at 4°C. Plates containing nutrient agar growth medium (Merck, Germany) were prepared with ethidium bromide concentrations from 0 to 2 mg/L. Strains were then cultured on nutrient agar plates containing different concentrations of ethidium bromide from the center of the medium to the side of the medium as a line. Concentrations depended on the type of bacteria and can be variable. The plates were incubated for 24 h at 37°C. Fluorescence of each isolate was measured with Gel Documentation (Hidolgh-Germany) to determine the lowest concentration of ethidium bromide affecting the bacteria (14, 15).
Phenotypic Evaluation of Efflux Pump Using Phenylalanine-arginine β-naphthylamide (Efflux Pump Inhibitor)
Phenylalanine-arginine β-naphthylamide dihydrochloride (Phe-Arg ß-naphthylamide dihydrochloride) was used to evaluate the phenotypic presence of efflux pump. First, two series of Mueller-Hinton agar growth medium (Merck-Elman) were prepared, the first series without inhibitory solution and the second series containing efflux pump inhibition solution. In Mueller-Hinton agar growth medium containing inhibitor, to dilute 0.25 mg phenylalanine β-naphthylamide dihydrochloride powder, 0.05 mg of this substance was dissolved in 30 mL of sterile distilled water and then 1 mL of this solution was transferred to empty sterile plates. Mueller-Hinton agar was then added to the plates (11) and multidrug-resistant M. catarrhalis isolates were cultured. Then standard discs, penicillin, ampicillin, amikacin, gentamicin, tetracycline, chloramphenicol, cefazolin, ceftazidime, trimethoprim-sulfamethoxazole, and ciprofloxacin disks were placed on the medium at standard intervals. The cultures were incubated at 37°C for 24 h and then the inhibited growth area was measured using a millimeter ruler. Measurements were classified as susceptible, resistant, semi-susceptible according to CLSI defined standards (13). The diameter of the inhibition zone was measured in two groups of plates containing phenylalanine-arginine β-naphthylamide and without phenylalanine-arginine β-naphthylamide. Increased diameter of the inhibition zone in the presence of the inhibitor indicates the effect of the efflux pump on antibiotic resistance. The results were analyzed using WHONET 5.6 software (WHO, Geneva Switzerland).
Frequency of Efflux Pump acrAB-oprM Gene in Multidrug-Resistant Isolates of M. Catarrhalis
PCR with specific primers was used to identify the efflux pump gene in multidrug-resistant isolates. Primers of efflux pump M. catarrhalis efflux pump genes were designed using the genes in NCBI and bioinformatics methods (Table 1).
Table 1. Designed primers used in the study.
| Gene | Primer | Gene length (Base pair) |
|---|---|---|
| 16s rRNA | F: 5 ́ CAGGCCTAACACATGCAAGTC3 ́ R: 5 ́ GGGCGGAGTGTACAAGGC3 ́ |
1360bp |
| oprm | F: 5 ́ GCCAGTCAAAAACAGCAAGC3 ́ R: 5 ́ TAATCCACCAATGCCGACTG3 ́ |
692 bp |
| acra | F: 5 ́ TTGGTTTAGAAGGCGGTGGC3 ́ R: 5 ́ TAGTATGGTGCAGGCAGGAC3 ́ |
1061bp |
| acrb | F: 5 ́ ACCACAGGTGAGGCAAGTAT3 ́ R: 5 ́ TGCCGATGGCGTTTGTTAAAT3 ́ |
719 bp |
At this point, Mastermix of Yekta Tajhiz (Iran) was used. The PCR mixture consisted of 18 μL of Mastermix, 1 μL of each of the forward and reverse primers, 2 μL of template DNA, and 3 μL of double distilled water with a total volume of 25 μL. The desired Mastermix was mixed with the primers of the efflux pump and the tubes containing the PCR mixture in a thermocycler (Sangavar-Bio-Rad) with a temperature program of 15 sec was initially denatured at 95°C and then 35 cycles including denaturing at 94°C for 5 sec, connection at 58°C for 15 sec, elongation at 72°C for 30 sec, and finally final elongation at 72°C for 120 sec were done. PCR products were then transferred to 2% agarose gel and identified by Gel Documentation electrophoresis (Hidolgh, Germany).
Data Analysis and Statistical Analysis
Data were evaluated using WHONET 5.6 software (WHO, Geneva Switzerland) according to CLSI (2016 version) and SPSS version 22 (SPSS Inc., Chicago, IL., USA). Chi-square test (chi-square) was used to analyze the relationship between the presence of efflux pump genes and drug resistance and an independent t-test was used to analyze the relationship between antibiotics and dependence on the efflux pump before and after using the pump inhibitor. 95% confidence level was considered for the significance of the tests (P≤0.05).
Results
After performing standard microbiological and biochemical tests, out of 137 randomly collected samples, including 80 men and 57 women aged 3-75 years, 10 isolates were identified as M. catarrhalis. The frequency of M. catarrhalis isolates by age and sex is shown in Table 2.
Table 2. Frequency of Moraxella catarrhalis isolates by age and sex.
Sample |
Age |
Sex |
Sample |
Age |
Sex |
| N1 |
65 |
Male |
N6 |
3 |
Male |
| N2 |
72 |
Male |
N7 |
72 |
Male |
| N3 |
69 |
Male |
N8 |
75 |
Male |
| N4 |
5 |
Female |
N9 |
45 |
Female |
| N5 |
66 |
Male |
N10 |
68 |
Male |
Frequency of M. catarrhalis isolates divided by clinical samples, 3 cases of sputum in patients with lower respiratory tract infection (30%), 1 case of smoker sputum with chronic bronchitis (10%), 2 cases of sputum in people with suspected tuberculosis (20%), as well as 2 cases of the adult hypopharynx and oropharynx swabs with sinusitis infection (20%) and 2 cases of purulent middle ear swabs in children (20%) were reported.
Results of Antibiotic Resistance by Disk Diffusion Method
According to the 2016 CLSI standard, out of 10 bacteria isolated from clinical samples, 7 isolates (70%) had the highest antibiotic resistance to the penicillin, ampicillin, amikacin, gentamicin, chloramphenicol, tetracycline, ciprofloxacin, cefazolin, ceftazidime and lower resistance to trimethoprim/sulfamethoxazole which showed to be resistant to more than two classes of antibiotics and were selected as MDR strains. All strains of bacteria were susceptible to amoxicillin/clavulanic acid (co-amoxiclav), azithromycin and erythromycin, and clarithromycin (100%). It was also found that none of the strains of M. catarrhalis are resistant to all antibiotics and the amoxicillin/clavulanic acid, clarithromycin, erythromycin, azithromycin antibiotics can be effective with 100% susceptibility.
Identification Results of Moraxella catarrhalis Isolated by PCR Test
After measuring the diameter of the inhibition zone, 7 isolates resistant to more than two classes of antibiotics were selected to perform the rest of the steps of this study. Using SrRNA16 specific primer, the isolates resistant to several drugs were identified. The formation of a band of 1360 pairs showed the presence of M. catarrhalis (Figure 1).
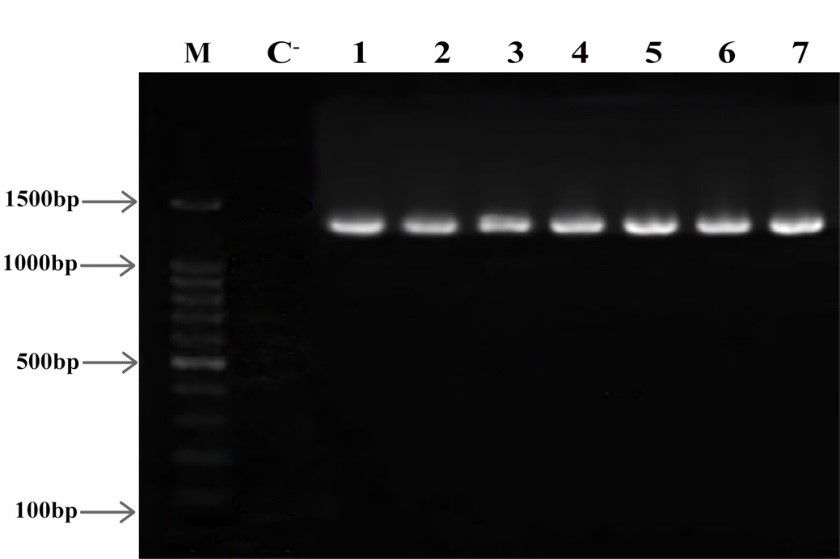
Figure 1. Electrophoresis of 16SrRNA gene product on 2% agarose gel M: Marker (Gene Ruler 100 bp) C-: Negative Control 1-7 columns: multidrug-resistant isolates Product Length: 1360 bp
Phenotypic Results of the Presence of Efflux Pump Using Ethidium Bromide and Cartwheel Method
To identify the efflux pump activity of M. catarrhal isolates, all isolates in the study were cultured with ethidium bromide agar technique and Cartwheel method. The results showed that 7 isolates (70%) had an efflux pump (also to ensure the presence of the Influx pump, all 10 isolates were evaluated). As shown in Figure 2, after being placed under a Gel Documentation device, 7 isolates of M. catarrhalis were unable to store these dyes in the cell.
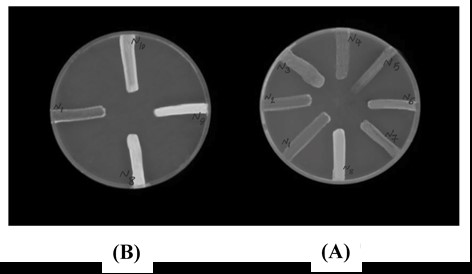
Figure 2. Results of phenotypic analysis of efflux pump. Each line is indicative of a Moraxella catarrhalis isolate. Hyaline lines are indicative of efflux pump inactivation and opalline lines are indicative of efflux pump activation. A: Fluorescence plate including 0.5 g/mL ethidium bromide and B: Fluorescence plate including 1 g/mL ethidium bromide
Results of the Dependence of Antibiotic Resistance on Efflux Pump Activity Using Efflux Pump Inhibitor
After the addition of the efflux pump inhibitor to the growth medium, cefazolin, ceftazidime, ciprofloxacin, chloramphenicol, tetracycline antibiotics became more antibacterial (Figure 3). These antibiotic resistance of M. catarrhalis isolates were dependent on the efflux pump because their growth inhibition zone changed compared to the first antibiogram (absence of inhibitor) and the diameter of the inhibition zone increased in the presence of inhibitor. Therefore, this issue shows the effect of the efflux pump on antibiotic resistance.
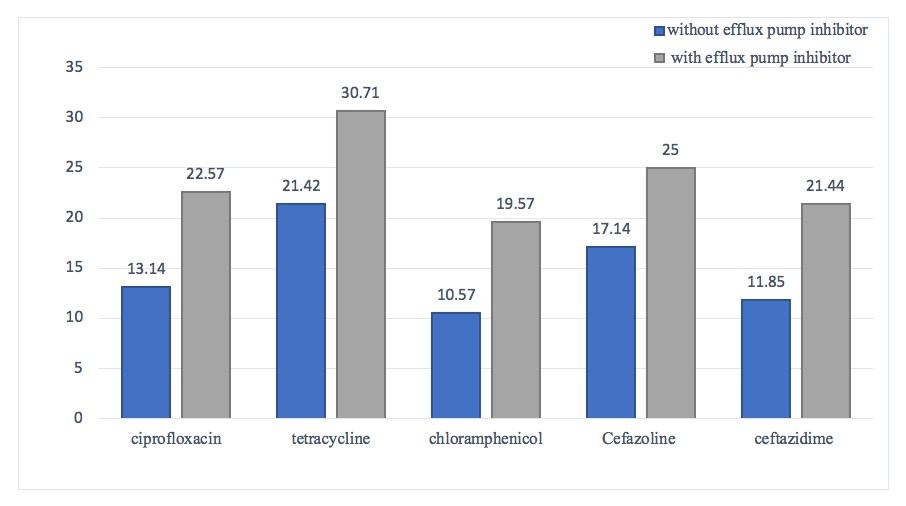
Figure 3. Comparing Moraxella catarrhalis isolates resistance patterns to ciprofloxacin, tetracycline, chloramphenicol, cefazoline, and ceftazidime with and without efflux pump inhibitor presence.
Results of Frequency Study of acra, acrb, oprm Genes
After amplification of acra, acrb, and oprm genes for all multidrug-resistant M. catarrhalis isolates, PCR products were observed in bands according to Figures 4, 5 and 6 on 2% agarose gel. The efflux pump genes were present in all 7 multidrug-resistant isolates (Figures 4, 5, 6).
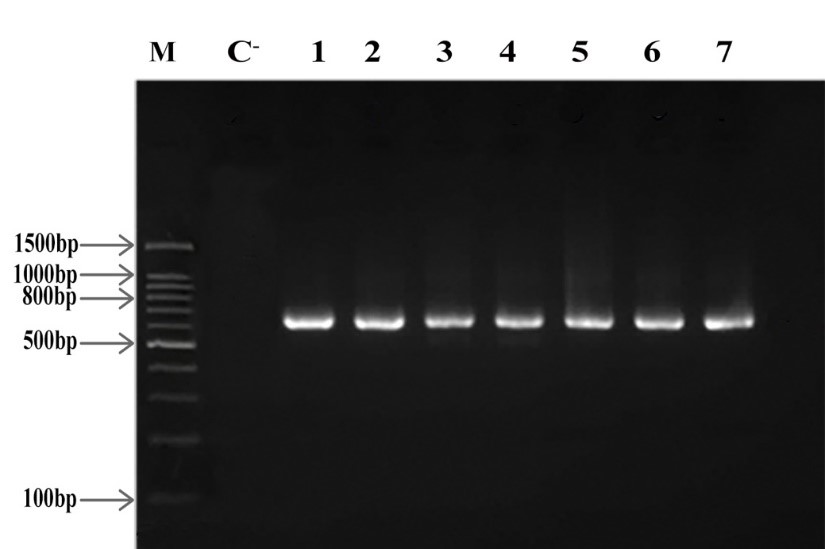
Figure 4. oprm electrophoresis on 2% agarose gel, M: Marker (Gene Ruler 100 bp), C-: Negative Control, 1-7 columns: multidrug-resistant isolates, Product Length: 692 bp

Figure 5. acrb electrophoresis on 2% agarose gel, M: Marker (Gene Ruler 100 bp), C-: Negative Control, 1-7 columns: multidrug-resistant isolates, Product Length: 719 bp
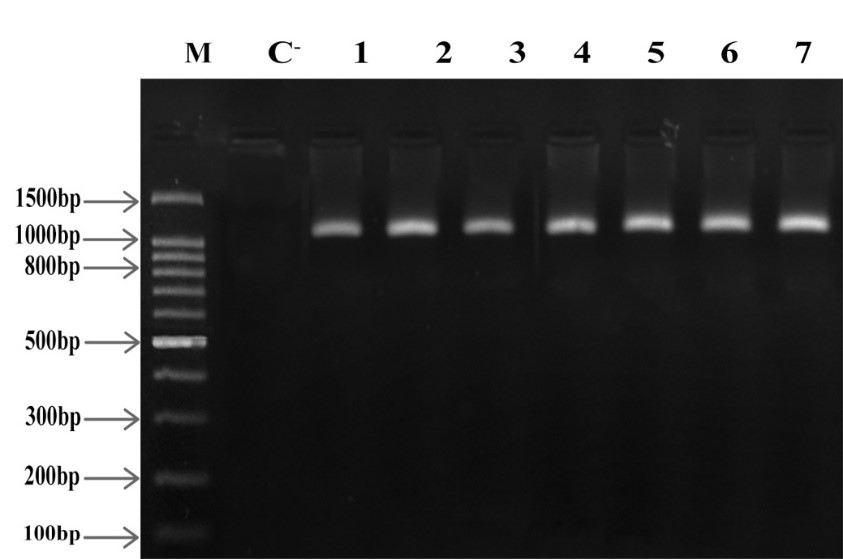
Figure 6. acra electrophoresis on 2% agarose gel, M: Marker (Gene Ruler 100 bp), C-: Negative Control, 1-7 columns: multidrug-resistant isolates, Product Length: 1061 bp
Also, according to the Chi-square test, a significant relationship was observed at the level of P-value≤0.05 between antibiotic-resistant isolates and the presence of acla, acrb and oprm genes in the efflux pump and no significant relationship was observed at the level of P-value≤0.05 for the four co-amoxiclav, erythromycin, azithromycin, and clarithromycin antibiotics, which were reported to be susceptible in all isolates.
Discussion
Moraxella catarrhalis has been reported in recent years as an important factor in lower and upper respiratory infections and it is the most common type that can be isolated from sputum, middle ear secretions, sinuses, and throat and mouth swabs (5). In this study, a total of 10 cases of M. catarrhalis were isolated from 137 different clinical samples. In a study conducted by Ghaznavi et al. (2005) on 200 patients, 17 samples of M. catarrhalis were isolated (16). A 2016 study by Sillanapau et al. on 222 specimens isolated 22 cases of M. catarrhalis (17). In a Miravitlles study of 48 patients with chronic obstructive pulmonary disease, eight M. catarrhalis patients (9%) isolated as the main cause of the disease (18). The mentioned values are somewhat consistent with the number of bacteria isolated in this study (10 samples). However, the small difference observed may be due to the different prevalence of bacteria in different geographical areas.
The results of antibiotic susceptibility showed that the highest resistance was related to gentamicin, amikacin, penicillin G, ampicillin, ceftazidime, cefazolin, tetracycline, chloramphenicol, and ciprofloxacin (70%). Isolates of this study showed moderate resistance to trimethoprim-sulfame-thoxazole antibiotics. Also, the isolated strains were completely susceptible to azithromycin, erythromycin, clarithromycin, and amoxicillin/ clavulanic acid antibiotics (100%).
Various studies have been performed to evaluate the antibiotic resistance of M. catarrhalis strains. In 2006, Naderi et al. isolated 54 samples of M. catarrhalis from 1161 children and examined the pattern of antibiotic resistance, with the highest susceptibility to amoxicillin/clavulanic acid (100%) and the highest resistance to penicillin (100%) (19). A 2014 study by Bandet et al. showed that all strains of isolated M. catarrhalis were highly susceptible to amoxicillin/clavulanic acid, erythromycin, and clarithromycin antibiotics (20). These results are consistent with the findings of the present study, which were 100% susceptible to amoxicillin/clavulanic acid and macrolides. Also, in a similar study conducted by Abdullah et al. in 2013 on 766 patients, 39 strains of M. catarrhalis were isolated, with the highest resistance to amikacin (92.3%) and the highest susceptibility to amoxicillin/clavulanic acid (100%). A study by Ramadan et al. in 2012 examined 200 patients, of whom 91.3% were resistant to penicillin and 100% susceptible to amoxicillin/clavulanic acid, ciprofloxacin, erythromycin, gentamicin, and 97.7% to tetracycline (22).
From the comparison of the present study and other studies published in the field of the prevalence of antibiotic-resistant strains of M. catarrhalis bacteria, it is clear that antibiotics such as penicillin, ampicillin, ceftazidime, cefazolin, chloramphenicol, tetracycline, ciprofloxacin, gentamicin, amikacin can no longer be used as a drug against M. catarrhalis due to repeated reports of resistance. One of the reasons for the increase in resistance in recent years is the excessive use of antibiotics, geographical and cultural factors such as the arbitrary use of antibiotics, and the availability of antibiotics, each of which has a role to play in increasing antibiotic resistance (23).
The AcrAB-OprM multidrug leakage system is the first pump under study in M. catarrhalis, which is a homolog of the efflux pump in Escherichia coli (5, 24). The number of studies performed on the efflux pump genes in M. catarrhalis is relatively limited and there are not many studies in this field. According to the results obtained in this study with phenotypic and genotypic tests, the presence of this pump and its involvement in antibiotic resistance were declared. The strength of this study was the phenotypic study at the same time as the genotypic study of the efflux pump activity, because despite the fact that the genotypic method is more accurate and reliable, simultaneous phenotypic and genotypic methods show more reliable results. One of the advantages of the phenotypic method is the faster cultivation and less expensive identification of the efflux pump (14, 15).
Also, the Cartwheel method, in addition to determining the phenotypic activity of the efflux pump, which shows the MDR phenotype in clinical isolates of gram-positive and gram-negative pathogenic bacteria; provides a rapid comparison of the activity of efflux resulting from isogenic mutations in the laboratory caused by continuous irradiation, deletion of a gene or group of genes, or growth of a bacterial strain under different conditions (temperature, pH, etc.) (14).
Based on the evidence, the activity of the efflux pump systems in M. catarrhalis was not previously fully identified. According to recent studies and PCR results of isolated strains in this study, it is shown that there are a number of efflux pumps in the M. catarrhalis bacteria and due to the increased resistance of M. catarrhalis and the increase in respiratory diseases, the study of these genes can be important in examining the treatment pattern of infectious and respiratory patients (5, 24). Using sequencing data for the BBH18 strain of M. catarrhalis in 2015, Spaniol et al. showed that Acr and Mtr efflux pumps were present. Also, following the previous studies, the authors (in 2010) showed that after treatment with amoxicillin, purine M35 is negatively regulated, which leads to increased resistance. These cascading reactions represent a new mechanism of resistance to aminopenicillins in M. catarrhalis. The role of AcrAB-OprM efflux pump in antibiotic resistance was also identified in that study by constructing mutant strains containing acrA, acrB, and oprM genes in M. catarrhalis O33E (5).
The phenylalanine-arginine-β-naphthylamide dihydrochloride inhibitor is one of the first inhibitors of RND pumps in gram-negative bacteria (26, 27). Studies show that by disrupting efflux pumps in resistant strains of bacteria with the help of efflux inhibitors, the properties of antibiotics and biocides are significantly increased (28). In a 2015 study by Gholami et al., examining 60 species of Acinetobacter baumannii, it was shown that susceptibility to imipenem increased in the presence of a phenylalanine-arginine-β-naphthylamide dihydrochloride inhibitor. So that for 96.6% of the isolates, the minimum inhibitory concentration decreased from 4 to 64 μg/mL (29). In 2013, Dal et al. examined the effect of PAßN and NMP inhibitors on the adeB gene in 40 Acinetobacter isolates. The antimicrobial properties of some antibiotics along with the efflux inhibitor increased significantly, but no significant change was reported in aminoglycosides (30). In 2010, Hornsey et al. showed a significant relationship in Acinetobacter between the expression of the efflux pump and the MIC (31). In a 2012 study by Mavri and Mozin, researchers found that disrupting efflux pumps in resistant strains of Campylobacter jejuni and Escherichia coli by efflux pump inhibitors increased the antimicrobial properties of antibiotics and biocides. The inhibitory effect of alpha naphthylamine was also investigated (32). The results of the above studies are consistent with the present study and show the importance of inhibiting the efflux pump.
In another 2012 study, Sonnet et al. showed that the phenylalanine inhibitor beta-naphthylamide and antibiotics such as ciprofloxacin in Pseudomonas aeruginosa were effective in suppressing resistance. For the clinical use of high-efficiency efflux pump inhibitor, a combination of inhibitors was prepared and their effect on different pumps was investigated (26). In the present study, it was shown that the efflux pump can be involved in reducing resistance to fluoroquinolones and some beta-lactams, tetracyclines, and chloramphenicol; Because by inactivating the pump, the effect of these antibiotics on all isolates increased. Therefore, the use of an efflux pump is an issue that can be considered to reduce the emergence of antibiotic-resistant species. There have been very few studies on the antibiotic resistance associated with the efflux pump and its inhibitor in the M. catarrhalis bacteria, but the results of this study were consistent with other bacteria. Given the increase in bacterial resistance and the significant role of efflux pumps in these resistances, the need to discover new antibiotics and inhibitor pump inhibitors seem obvious (33). Although antibiotic resistance may not completely restore susceptibility to multidrug-resistant organisms only by inhibiting efflux pumps, given the maximal activity of the efflux pump in 70% of the strains of this study, the association between the expression of efflux pumps and antibiotic resistance should not be overlooked. Genetic studies of these pumps and other involved factors including beta-lactamase enzymes will be considered necessary in future studies.
Conclusion
According to the approval of phenotypic and genotypic role of efflux pumps and dependence of antibiotic resistance to efflux pump activity using pump inhibitors, research and novel drugs can be developed in order to control and treat Moraxella catarrhalis infections. Development of efflux pump inhibitors can revive antibiotic efficacy. On the other hand, new proposals can be practiced to inhibit efflux pumps in order to decrease drug resistance in M. catarrhalis.
Acknowledgements
I would like to express my gratitude and thanks to all those who helped me during the implementation of this research. It is worth mentioning that this article is the result of a part of the master's thesis of Ms. Parvin Mohammad Shafiei from the Islamic Azad University, Kazerun Branch.
Conflicts of Interest
Authors declared no conflict of interests.
Financial Resources
This article was financially supported by the authors.
Received: 2020/06/3 | Accepted: 2020/08/10 | ePublished: 2020/09/1
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |








