BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-996-en.html
2- Department of Microbiology, College of Medicine, University of Karbala, Karbala, Iraq , aljanabibio@gmail.com
.
Dermatophytes are a specialized group of filamentous fungi living in an environment enriched with keratinized materials. They have an ability to invade the skin, hair, and nail causing cutaneous diseases in the human called dermatophytosis (1). Dermatophytes like other filamentous fungi can grow and increase their number by sexual and asexual reproduction (2). The asexual process is the main type of multiplication that is followed by almost all species of dermatophytes for reproduction, while few of them are known to have sexual stages (3). Production of conidia, which can mainly take microconidia and macroconidia forms as a final product, is an important asexual stage in dermatophytes (2).
Development of asexual reproduction in some filamentous fungi has proved to be under the regulation of three genes called brlA, abaA, and wetA (4, 5). Inactivation of these genes causes major abnormalities in conidia production and conidiophore morphology without affecting vegetative growth (4, 5). For regulation of conidia production, these genes usually work in a cascade process starting with activation of brlA gene, which triggers the expression of the other two genes (4). Thus, any inactivation of brlA will significantly affect the conidiation roles of the abaA and wetA genes and any excessive expression of these genes will induce cessation of vegetative growth and cellular vacuolization from the beginning of conidiophores
vesicle formation (4, 5). However, the expression pattern of such genes is associated with the level of RNA accumulation through their activity on codons of poly(A)+RNAs that begin to accumulate at different times during the conidiation process (4).
Genomes of dermatophytes are recognized by highly collinear and similar in size, ranging from 22.5 Mb for Trichophyton rubrum to 24.1 Mb for T. equinum (6). Thus, the growth of a dermatophyte usually needs approximately 2044 differentially expressed genes (7). Among this huge number of genes, brlA, abaA, and wetA may have a role in the development of asexual conidia of dermatophyte. This may be cleared when the presence of these genes can firstly prove in dermatophytes, which is the main goal of this study.
Three genera with seven strains of dermatophytes with an identification barcode No. in GenBank were obtained from the College of Medicine, University of Karbala. They comprised one strain of T. interdigitale (MH383047), one of Epidermophyton floccosum (MH383041), two strains of Microsporum ferrugineum (MH383042 and MH383043), and three strains of Microsporum canis (MH383044, MH383045, and MH383046). Fungi were activated by culturing on Sabouraudʼs Dextrose Agar [(SDA), HiMedia, India] for one week at 28-30°C.
Molecular Diagnosis Method
The DNA of dermatophytic strains was extracted by using FavorPrepTM Fungi/Yeast Genomic DNA Extraction Mini Kit (Favorgen Biotech Corp., Taiwan). Primers of brlA, abaA, and wetA genes were designed by Primer3 web designer (version 4.1.0) (http://primer3.ut.ee/) based on gene sequences of three fungi registered in GenBank (Table 1). Primer of brlA was designed based on gene sequences of Aspergillus nidulans under the code M20631.1. Primer of wetA was designed based on gene sequences of Arthroderma otae CBS 113480 with a barcode NCBI reference sequence: XM_002850912.1. Primer of abaA was designed based on gene sequences of Emericella nidulans with a barcode J04850.1.
PCR mixture was set up in a total volume of 20 μl, including 5μl of PCR master mix of AccuPower® PCR Premix (Bioneer, South Korea), 1μl of each primer (10 µM), 1μl of template DNA (100 ng) and sterile deionized distilled water. Negative control containing all material except template DNA was also used.
Table 1. Primers of brlA, abaA, and wetA genes (20)
| Gene name | Size (bp) | Primer |
| brlA | 155 | F- tcatcaagcaggtgcagttc |
| R- gtcactccgtgagaaggctc | ||
| abaA | 138 | F- agctatcttgcgcaccctaa |
| R- acgaagaaaggcagacagga | ||
| wetA | 207 | F- cgaaatgctccaaaccaaat |
| R- ttctcttgtcacgggcttct |
Table 2. Conidiation genes in dermatophytes
| Dermatophyte name | No. in genbank | ablA gene | abaA gene | wetA gene |
| E. floccosum | MH383041 | - | - | - |
| M. ferrugineum | MH383042 | - | - | - |
| T. interdigitale | MH383047 | - | + | - |
| M. ferrugineum | MH383043 | + | + | + |
| M. canis | MH383044 | + | + | + |
| M. canis | MH383045 | + | + | + |
| M. canis | MH383046 | + | + | + |
1: E. floccosum, 2: M. ferrugineum MH383042, 3: T. interdigitale, 4: M. ferrugineum MH383043, 5: M. canis MH383044, 6: M. canis MH383045, 7: M. canis MH383046, 8: negative sample
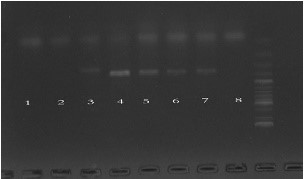
1: E. floccosum, 2: M. ferrugineum MH383042, 3: T. interdigitale, 4: M. ferrugineum MH383043, 5: M. canis MH383044, 6: M. canis MH383045, 7: M. canis MH383046, 8: negative sample
Production of conidia is an important process performed by fungi to ensure its continuous presence on the Earth. This type of multiplication process is usually regulated by the expression of several genes. Studies with Aspergillus nidulans revealed the presence of three genes, brlA, abaA, and wetA, which were considered later as a central regulatory pathway of conidia production and conidophore in filamentous fungi (4). Regulatory effects of such genes either on their own expression or on other sporulation-specific genes could be performed individually or together (5).
From the results of the current study, the contents of regulatory genes involved in conidiation development were observed to show variation among different dermatophyte strains. All strains of M. canis, which is one of a zoophilic type of dermatophytes (2), were shown to have all of the investigated genes. Detection of such specific genes in dermatophytes is assumed to be not easy due to the genome content of gene families in this fungal group, which mostly differed from other human-associated fungi (6). Otherwise, thousands of genes, which are responsible for several growth processes were identified in dermatophyte as in T. rubrum and only 37 homologous genes from this content are specifically involved in several signaling pathways or regulation modules (7). Conidiation genes in filamentous fungi usually work together in a activation cascade to regulate the sporulation process (5). brlA is an important gene for conidiation, which encodes a 432 amino acid polypeptide containing two directly repeated motifs resembling the Zn (II) coordination sites (8). Activation of brlA, which leads to synthesis of BrlA polypeptide, is considered as the first essential step in the progression of the cascade genetic regulatory pathway of conidiation (9). This activation not only affected asexual spore development, but also the sexual process (10). The brlA of A. nidulans is consist of two overlapping transcription units, alpha and beta, which are essential to form morphologically normal conidiophores (11). However, any overexpression of brlA gene in vegetative cells will lead to growth cessation, formation of viable conidia on the hyphal tips and direct production of phialides without needing to form vesicle and metula first (9).
The second detected gene in dermatophytes is abaA gene, which is usually activated by the action of brlA, especially during the middle stage of the conidiation or condiophore development after differentiation of metulae as noted in A. fumigatus (9, 12). This gene is predicted to encode an ATTS/TEA DNA-binding domain protein and it could play as a feedback regulatory factor for conidiophore development through the action of multiple structural binding sites in its encoded protein AbaA (12). In Penicillium marneffei, abaA was found to regulate the events of the cell cycle and dimorphic transformation (13). However, analysis of RNA sequences showed that AbaA protein in Fusarium graminearum plays an important role in cell cycle pathways and other conidiation-related genes (14). Deletion of abaA gene results in production of aberrant conidiophores in the form of reiterated cylinder-like terminal cells lacking spores and causes delayed autolysis and cell death (12).
The wetA gene is the third regulatory gene of the conidiation process investigated in the genomic content of dermatophytes in this study. The wetA gene activity is found to be affected by abaA only and not brlA (9). An activation of wetA usually leads to control of spore maturation at the late stage of conidiation process through directly or indirectly regulated expression of some spore-specific genes (12). It is specifically required for the synthesis of cell wall and its crucial form (12). wetA is found to encode a polypeptide enrichment with serine (14%), threonine (7%), and proline (10%) (5).
T. interdigitale frequently causes different types of dermatophytosis on the human skin such as tinea pedis, tinea corporis and onychomycosis, which can produce a variable texture of colonies on culture media (15). The strain used in this study had abaA gene but no other two genes. Such type of dermatophyte, which is related to the T. mentagrophytes complex, conidia is most frequently absent, or it produces very few of them (16). In production state, macroconidia often take spherical or subspherical shapes, while macroconidia are rarely observed (15, 16). Absence of ablA and wetA genes could play a role in reduction of conidiation development in the currently used strain of T. interdigitale, but this is not the ultimate decision and needs more research to confirm.
The presence of conidiation genes in M. ferrugineum strains showed variable results. Strain No. MH383043 revealed has all of these genes, while they absolutely absent in strain No. MH383042. This anthropophilic dermatophyte, which is usually caused tinea capitis in most cases of dermatophytosis, rarely formed conidia in normal conditions (17). Thus, diagnosis of such type of dermatophytes is preferred using PCR technique, which gives an excellent sensitivity (100%) (17). A few strains were found to have an ability to produce microconidia and/or macroconidia on Casero medium (18). Therefore, detection of conidiation genes in such type of dermatophytes could be observed to vary depending on strain type.
As M. ferrugineum MH383042, E. floccosum showed no content of selective conidiation genes. Such species, which is frequently associated with other dermatophytes to cause tinea cruris and tinea pedis, normally produces abundant club-shaped, smooth-walled, 2 to 4 celled macroconidia without microconidia (19). Thus, conidiation process in E. floccosum may be regulated by other genes not found in other types of the human pathogenic fungi (6).
brlA, abaA, and wetA genes in dermatophytes is variable and strain dependent. M. canis is the species with highest content of conidiation genes, while T. interdigitale and E. floccosum lacked such genes. The conidiation process in dermatophytes is assumed to be under the control of other genes not included in this study. More experiments are needed to explain the main role of brlA, abaA, and wetA genes in conidiation process of dermatophytes and this study could be the first step.
The authors thank all those who helped them writing this article.
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
This research resulted from an independent research without receiving any financial support.
Authors declared no conflict of interests.
- Vander Straten MR, Hossain MA, Ghannoum MA. Cutaneous infections dermatophytosis, onychomycosis, and tinea versicolor. Infect Dis Clin North Am. 2003;17(1):87-112. [DOI:10.1016/S0891-5520(02)00065-X]
- Weitzman I, Summerbell RC. The dermatophytes. Clin Microbiol Rev. 1995;8(2):240-59. [DOI:10.1128/CMR.8.2.240] [PMID] [PMCID]
- Metin B, Heitman J. Sexual Reproduction in Dermatophytes. Mycopathologia. 2017;182(1-2):45-55. [DOI:10.1007/s11046-016-0072-x] [PMID] [PMCID]
- Boylan MT, Mirabito PM, Willett CE, Zimmerman CR, Timberlake WE. Isolation and physical characterization of three essential conidiation genes from Aspergillus nidulans. Mol Cell Biol. 1987;7(9):3113-8. [DOI:10.1128/MCB.7.9.3113] [PMID] [PMCID]
- Adams TH, Wieser JK, Yu J-H. Asexual Sporulation in Aspergillus nidulans. Microbiol Mol Biol Rev. 1998;62(1):35-54. [DOI:10.1128/MMBR.62.1.35-54.1998] [PMID] [PMCID]
- Martinez DA, Oliver BG, Graser Y, Goldberg JM, Li W, Martinez-Rossi NM, et al. Comparative genome analysis of Trichophyton rubrum and related dermatophytes reveals candidate genes involved in infection. mBio. 2012;3(5):e00259-12. [DOI:10.1128/mBio.00259-12] [PMID] [PMCID]
- Xu X, Liu T, Leng W, Dong J, Xue Y, Yang H, et al. Global gene expression profiles for the growth phases of Trichophyton rubrum. Sci China Life Sci. 2011;54(7):675-82. [DOI:10.1007/s11427-011-4187-5] [PMID]
- Adams TH, Boylan MT, Timberlake WE. brlA is necessary and sufficient to direct conidiophore development in aspergillus nidulans. Cell. 1988;54(3):353-62. [DOI:10.1016/0092-8674(88)90198-5]
- Park HS, Yu JH. Genetic control of asexual sporulation in filamentous fungi. Curr Opin Microbiol. 2012;15(6):669-77. [DOI:10.1016/j.mib.2012.09.006] [PMID]
- Tsitsigiannis DI, Kowieski TM, Zarnowski R, Keller NP. Three putative oxylipin biosynthetic genes integrate sexual and asexual development in Aspergillus nidulans. Microbiology (Reading). 2005;151(Pt 6):1809-21. [DOI:10.1099/mic.0.27880-0] [PMID]
- Drira I, Hadrich I, Neji S, Mahfouth N, Trabelsi H, Sellami H, et al. Highly discriminatory variable-number tandem-repeat markers for genotyping of Trichophyton interdigitale strains. J Clin Microbiol. 2014;52(9):3290-6. [DOI:10.1128/JCM.00828-14] [PMID] [PMCID]
- Tao L, Yu JH. AbaA and WetA govern distinct stages of Aspergillus fumigatus development. Microbiology (Reading). 2011;157(Pt 2):313-26. [DOI:10.1099/mic.0.044271-0] [PMID]
- Borneman AR, Hynes MJ, Andrianopoulos A. The abaA homologue of Penicillium marneffei participates in two developmental programmes: conidiation and dimorphic growth. Mol Microbiol. 2000;38(5):1034-47. [DOI:10.1046/j.1365-2958.2000.02202.x] [PMID]
- Son H, Kim MG, Min K, Seo YS, Lim JY, Choi GJ, et al. AbaA regulates conidiogenesis in the ascomycete fungus Fusarium graminearum. PLoS One. 2013;8(9):e72915. [DOI:10.1371/journal.pone.0072915] [PMID] [PMCID]
- Prade RA, Timberlake WE. The Aspergillus nidulans brlA regulatory locus consists of overlapping transcription units that are individually required for conidiophore development. The EMBO Journal. 1993;12(6):2439-47. [DOI:10.1002/j.1460-2075.1993.tb05898.x]
- de Hoog GS, Guarro J, Gené J, Figueras M. Atlas of clinical fungi: Centraalbureau voor Schimmelcultures (CBS); 2000.
- Yuksel T, Ilkit M. Identification of rare macroconidia-producing dermatophytic fungi by real-time PCR. Med Mycol. 2012;50(4):346-52. [DOI:10.3109/13693786.2011.610036] [PMID]
- Weitzman I, Rosenthal S. Studies in the differentiation between Microsporum ferrugineum Ota and Trichophyton soudanense Joyeux. Mycopathologia. 1984;84(2-3):95-101. [DOI:10.1007/BF00436519] [PMID]
- AL-Janabi A. Study of Characteristic Features of Pleomorphic Epidermophyton floccosum. Glob Environ Res. 2009;3(2):132-4.
- Katz ME, Braunberger K, Yi G, Cooper S, Nonhebel HM, Gondro C. A p53-like transcription factor similar to Ndt80 controls the response to nutrient stress in the filamentous fungus, Aspergillus nidulans. F1000Research. 2013;2. [DOI:f1000research.2-72.v1] [PMID]
Received: 2019/11/19 | Accepted: 2020/09/20 | ePublished: 2020/10/27
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |









