BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1776-en.html
2- Department of Microbiology, Science Faculty, Arak Branch, Islamic Azad University, Arak, Iran , jafarizohreh264@gmail.com
Shigellosis or basil bloody diarrhea is the second leading cause of death in children under the age of 5, especially in developed and developing countries. The cause of this disease is a bacterium called Shigella. Shigella are facultative anaerobic gram-negative bacilli, non-motile and without spores, which have 4 subgroups: Shigella dysenteriae, Shigella flexneri, Shigella boydii, and Shigella sonnei (1, 2). Shigellosis with low infectious dose (10-100 bacteria), the emergence of antibiotic resistance, complications, and high mortality have been major global health problems. Statistics show that Shigella is the cause of 165 million bloody diarrhea that occurs per year, so prevention and timely treatment of this disease is significant (3, 4). The most important way to get it into the body is fecal-oral. Due to the low-speed infectious dose, its transmission from person to person is very high so this disease can spread in communities with health poverty (5-7). The most important damage of this bacterium is its invasion of mucosal epithelial cells (M cells) which causes damage to the coating tissue and ulcers in the intestinal mucosa. Finally, with the loss of blood and release of inflammatory elements and mucus, water absorption is prevented, the volume of feces changes and bloody and mucous diarrhea occurs (8, 9). The rate of increase of antimicrobial resistance in Shigella isolated from humans has become important during the last two decades.
Recent studies show that Shigella can acquire antibiotic-resistance genes, including aminoglycosides, through plasmids and integrons (10). Aminoglycosides are a group of antibiotics that bind to the 30s component of bacterial ribosomes to inhibit the synthesis of bacterial proteins. Ultimately, it reduces or eliminates the power of pathogenicity. The clinical significance of these antibiotics is that they positively affect a wide range of facultative anaerobic bacteria such as gram-negative, staphylococci, and streptococci, and are used in bacilli infections such as Shigella. Shigella contain aminoglycoside-modifying enzymes (AMEs), and these enzymes cause resistance through three reactions: Adenylation, Phosphorylation, and Acetylation. Recent studies have shown that aadA1 and aadA2 genes are present in Enterobacteriaceae, especially Shigella, which cause resistance to streptomycin and spectinomycin (11, 12). There are strains of Campylobacter jejuni that have the aph3-1 and aadE genes that make them resistant to aminoglycosides. The aadE gene induces resistance to streptomycin (13).
Also, the aph gene has been shown to cause resistance to neomycin and kanamycin in gram-negative gut bacteria. The aac gene is one of the most important aminoglycoside resistance genes that cause gentamicin resistance in gram-negative bacteria such as Shigella. Unfortunately, gentamicin resistance is widespread in these strains today (14). Curcumin is a polyphenolic compound with the chemical name Diferuloylmethane and the chemical formula C21H20O6, extracted from turmeric and widely used in the food industry. Turmeric root contains 2-9 % of curcumin. For centuries, Asian countries have used turmeric as an antibacterial, anti-inflammatory and anti-cancer agent in their traditional medicine. Curcumin exerts its therapeutic effects by interacting with biological molecules such as tumor cytokines, transcription factors, kinases, enzymes, receptors, growth factors, and proteins (15, 16). The possible mechanisms of action of curcumin that scientists have found are that initially, by forming an ion channel, it causes pores in the outer membrane and the cell wall. With the outflow of important metabolites of the bacterial cell, it eventually leads to the death of the bacterium. In addition, the bacteriostatic function of curcumin by inhibiting the bacteria's DNA repair process is also considered an antibacterial mechanism of curcumin (17, 18). Curcumin has been reported to inhibit the growth of various Shigella species in appropriate doses (19). Therefore, the aim of this study is to investigate the molecular resistance genes to the aminoglycoside of S. dysenteriae isolated from children and the expression of aadE under the influence of curcumin nanoparticles.
Sampling and Isolation
In this cross-sectional study, 60 stool samples were collected randomly from children less than ten years old with diarrhea referred to Tehran hospitals, Iran, in 2022. All samples were transferred to Pasargad Laboratory (Tehran, Iran). The samples were first cultured on MacConkey agar and then Salmonella-Shiglla agar (Merck Co. Germany) and incubated at 37°C for 24h. After identifying and confirming the presence of Shigella bacteria, differential tests of TSI (Triple sugar iron agar) (Merck Co., Germany), IMViC, urease and decarboxylation of amino acids ornithine, and mannitol were conducted for the final diagnosis of S. dysenteriae species (1, 19).
Antibiogram Test
Antibiotic sensitivity test using the disk diffusion method and based on the instructions of the Laboratory and Clinical Standards Institute (1977), on Mueller Hinton Agar medium (Merck, Germany) was conducted for gentamicin (10μg), kanamycin (30μg), vancomycin (30μg), Tobramycin (30μg), amikacin (30μg) antibiotics (manufactured by Himedia, India).
DNA Extraction and Multiplex PCR Reaction
After bacterial DNA extraction using DNA Cinnagen kit and confirmation of the extracted DNA purity using a biophotometer (Bio-Rad, USA), The aadE, aacA-aphD, and aph genes were amplified BY using Multiplex PCR method and specific primer sequences (Table 1) in a thermocycler (Eppendorf, Germany) in the final volume of 20 µL including 10 µL PCR master mix 2X (Sinaclon, Iran) containing Taq DNA polymerase (0.05 U/µL), MgCl2(3mM) and dNTPs (10mM), 1 μL of each primer, 4 μL of template DNA (40 ng) and 4 μL of double-distilled water for 33 cycles. Thermal cycling included a denaturation step at 95°C for 30 sec, primer annealing at 30°C for 55 sec, and an elongation step at 72°C for 60 s. Finally, PCR products were run on 1% agarose gel.
Table 1. Primers used in Multiplex-PCR (20-22)
| Size | Primer sequence | GEN |
| 198bp | GCCCTTGGAAGAGTTAGATAATT CGGCACAATCCTTTAATAACA |
aadE |
| 220bp | CCAAGAGCAATAAGGGCATA CACTATCATAACCACTACCC |
aacA- aphD |
| 125bp | GAGGGCTTTAGGAATTACGC ACACACCGACCAACAGAGG |
aph |
Preparation of Shigella dysenteriae Microbial Suspension with a Concentration Equivalent to 0.5 McFarland
The reason for preparing 0.5 McFarland is that in the sense of standard inoculation for microbial tests, standard barium sulfate equal to half-McFarland standard should be used. For example, a 0.5 McFarland tube contained an approximate density of bacteria. To prepare this standard, 0.05 mL of barium chloride 1% was mixed with 9.95 mL of sulfuric acid 1%. Bacterial culture is required for 24 hours to prepare the microbial suspension. Therefore, 24 hours before the experiment, the culture is inoculated with nutrient agar in a sloping medium and incubated for 24 hours at 37°C. A 24-hour bacterial culture was used and dissolved in normal saline. The suspension was returned at 530 nm with the absorption of 0.5 McFarland solution. In other words, the produced suspension should contain 1.5 × 108 (21).
Preparation of Curcumin Nanoparticle Suspension
All nanoparticle formulations were measured in the range of 2–40 nm as determined by electron microscopy (23). To prepare the nanoparticle stock solution, 10 grams of nanoparticles were suspended in one liter of 50% dimethyl sulfoxide (DMSO) as an auxiliary solvent, and the Ultrasonic RK 31 H was used for 30 minutes to properly disperse them (24, 25).
Determination of MIC Nano Curcumin
This method used a 96-cell micro-plate with 12 rows containing 100 μL of sterile Mueller Hinton Broth medium. Then 100 μL of high dilution nanoparticles of 1024μg / mL curcumin was added to the first row containing 100 μL of Mueller Hinton Broth, and dilution was done until 128 dilutions (128-1024 μg / mL). Next, 100 μL of half-McFarland microbial suspension was added to all wells. Also, two rows of wells were used as the positive control (environment and microbial suspension) and negative control (environment + curcumin nanoparticles) of the test. Then the microplates were incubated in a shaker incubator (200 rpm, 37°C for a maximum of 24 hours) (26). Next, we will investigate the expression of aadE gene in the presence of curcumin nanoparticles.
Real-time PCR Test
RNA was extracted by Qiagen RNeasy microkit from strains carrying the aadE gene based on the Sub MIC and considering both treated and untreated groups in the logarithmic growth phase (at 0.4-0.6 = OD600). Also, Qiagen DNase kit was used to remove DNA. The accuracy of RNA extraction was assessed by spectrophotometry. cDNA was synthesized using Roche's Reverse AMV enzyme. The required volume for a Real-Time polymerase chain reaction is 20 µL, and this reaction is conducted using the Genet bio CAT kit. NO: Q9210 South Korea was done as follows: 10 μL of Primer Qmaster mix (2x) with syber green, 5 μL of Depc water, 1 μL of Primer Forward, 1 μL of Primer Reverse, 1 μL of Rox color, and 2 μL of cDNA was used. Amplification of the desired parts was done in the Corbet machine with an initial denaturation program of 95°C for 1 minute / 95°C for 30 seconds / 59°C for 40 seconds / 72°C for 60 seconds, in 35 cycles. The 16 SrRNA home gene was used as an internal control of the test (26).
Cultivation, Isolation, and Identity of Bacteria
A total of 60 stool samples suspected of Shigella dysenteriae were collected from 4 to 10-year-old children with diarrhea referred to Tehran Hospitals, in Pasargad Research Laboratory (Tehran, Iran). Colorless colonies on McConkey agar and Salmonella-Shigella agar were considered positive samples. Among the analyzed samples, 12 isolates belonged to the Shigella dysenteriae strain, which had the biochemical characteristics of lactose negative, gas negative, indole negative, non-motion, ornithine and mannitol decarboxylation reaction negative, urease negative, and citrate negative, methyl red positive and Voges-Proskauer negative.
Sensitivity Test
8 isolates showed resistance to gentamicin and tobramycin, 1 isolate to amikacin, 1 isolate to kanamycin, and no resistance was observed against vancomycin. No resistance was observed in 2 isolates.
Multiplex PCR Reaction
From 12 Shigella dysenteriae isolates identified according to Figure 1, 10 isolates (83.3%) carried the aadE gene, 6 isolates carried the aacA_aphD gene, and 1 isolate carried the aph gene.
MIC Test
The results of the MIC of curcumin nanoparticles in Shigella dysenteriae showed that the minimum concentration of growth inhibition in the presence of curcumin nanoparticles was 225 μg/mL and the Sub MIC of curcumin nanoparticles to reduce aadE gene expression was 128 μg/mL.
aadE Gene Expression after Exposure to Curcumin Nanoparticles
Due to the greater abundance of aadE gene, the expression of this gene was measured under the influence of curcumin nanoparticles, and by using the Real-time PCR technique, the reproduction curve is shown in Figure 2. The relative expression of aadE gene in isolates treated with Curcumin nanoparticles showed a significant change compared to the isolates untreated and based on the results, the P-value for aadE gene is less than 0.05, which indicates the significance of the difference in the expression of these two genes between the treated and untreated groups. The Fold Change parameter for the aadE gene is -1.03, indicating that this gene was reduced by 1.03 times in the treated group compared to the untreated group.
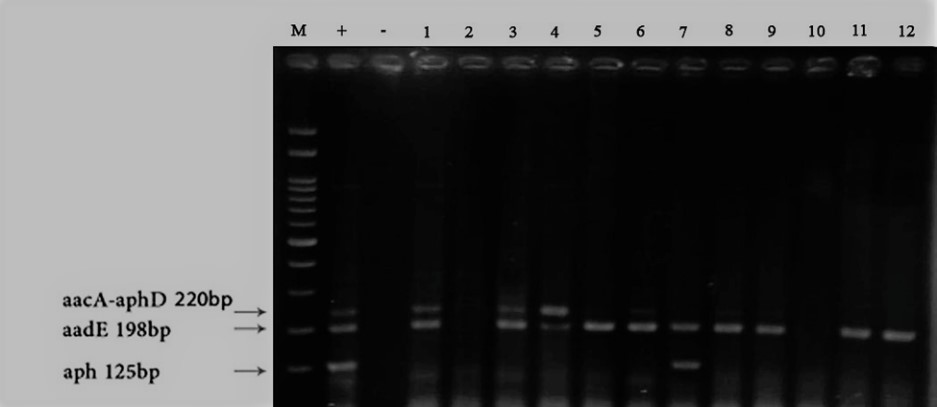
Figure 1. Results of Multiplex PCR and electrophoresis on some positive isolates from samples 1 to 12, 100 bp marker, +: positive control, and -: negative control.

Figure 2. Results of Real-Time PCR reproduction curve of aadE gene related to aminoglycoside resistance.
Shigellosis or basil bloody diarrhea is important due to the prevalence of bacteria strains resistant to aminoglycosides and high mortality in children under 5, so early diagnosis and antibiotic treatment of shigellosis is very important and greatly reduces the complications caused by it. Antibiotic-resistant strains such as aminoglycosides have been reported worldwide. Studies have shown that Shigella strains contain aminoglycoside-modifying enzymes (AMEs) that induce resistance to aminoglycosides through three reactions: adenylation, phosphorylation, and acetylation (11). Studies have shown an increase in the prevalence of aph, aacA-aphD, and aadE genes among bacteria. So Vaziri et al. Showed that aac (6ʹ) -Ⅰ, aac (6ʹ) -Ⅱ, and ant (2ʺ) -Ⅰc and aph(3ʹ)- Ⅳ genes in Pseudomonas aeruginosa cause aminoglycoside resistance in Iran, and in this study aac(6ʹ)-Ⅱ gene by 36% was the most common gene and the lowest gene was aph(3ʹ)- Ⅳ (27).
In our study, the amount of aph gene was very low and was observed in only one strain of Shigella dysenteriae. Also, in 2019, Silva et al. showed that Campylobacter jejuni isolates contain aph3-1 and aadE genes, which cause aminoglycoside resistance. On the other hand, aph3-1 resistance gene was the most common gene, and aadE gene had the lowest abundance among Campylobacter jejuni isolates, and strains with aadE gene showed resistance to streptomycin. These results are not consistent with our study because the aadE gene was the most common gene (83.3%) among Shigella dysenteriae isolates (13). In general, it can be concluded that this gene causes streptomycin resistance in Shigella dysenteriae and Campylobacter jejuni. In 2003, Strommenger et al. in a study showed that the presence of aacA-aphD1 and aacA-aphD2 genes in Staphylococcus aureus makes strains resistant to gentamicin, and about half of the strains contained these genes, which was a significant amount (28). The results of this study were consistent with our research because the aacA-aphD gene was also seen in half of Shigella dysenteriae strains, so it can be concluded that the aacA-aphD aminoglycoside resistance gene is present in both gram-positive and gram-negative bacteria. The results of Moniri et al.'s research in 2010 showed that out of 60 isolates of Acinetobacter baumannii examined, more than 60% contain aphA6 and aac1 genes, which cause resistance to amikacin and tobramycin (29). The results of this study were consistent with our study because the abundance of aacA-aphD gene was 50%. In other words, it can be concluded that the aac gene in gram-negative bacteria causes resistance to aminoglycosides and that this gene should be considered. Turmeric has long been regarded for its antibacterial, therapeutic, and anti-cancer properties, and people in Asian countries have used this valuable substance to treat inflammations, gastrointestinal and respiratory diseases, and toothache. The pharmacological and antibacterial properties of turmeric are due to the presence of an important phenolic compound called curcumin. This property of curcumin is due to the presence of hydroxyl groups in the aromatic ring of phenolic compounds. At higher concentrations of phenolic compounds, due to the increase in the number of hydroxyl groups, the probability of hydrogen donation to the free radical and, following that, inhibition increases (30). Curcumin is less used in clinical cases due to its bioavailability and low absorption in the body, and scientists have improved its effectiveness with nanotechnology (17).
Now, with the advent of aminoglycoside-resistant strains, the question is whether herbal medicines such as curcumin, despite their pharmacological and antibacterial properties, can reduce the expression of aminoglycoside resistance genes in Shigella dysenteriae bacteria, the cause of shigellosis. Therefore, our study is about the effect of curcumin nanoparticles on the expression of aadE gene, which was present in 83.3% of the isolates. So far, studies on the antibacterial effects of curcumin on bacteria have been conducted. For example, De et al. showed that the growth of 81% of Helicobacter pylori isolates was inhibited at a concentration of 10 μg/mL of curcumin (31). The MIC results of this study were inconsistent with our study, but the growth-inhibiting effect of curcumin on gastrointestinal bacteria is proven. Shariati et al. showed that the MIC of water-soluble curcumin nanoparticles (128 μg/mL) was less than that of nanoparticles dissolved in dimethyl sulfoxide (256 μg/mL) for Pseudomonas aeruginosa (32). In our study, curcumin nanoparticles were dissolved in dimethyl sulfoxide, and their MIC was almost identical to the MIC of nanoparticles dissolved in De's study. In a research conducted by Kareem et al. to study curcumin on Shigella dysenteriae and Campylobacter jejuni in 2020, the MIC of curcumin for Shigella dysenteriae was 256 µg/mL and for Campylobacter jejuni was 512 µg/mL (19). In our study, nanotechnology was used to improve the effectiveness of curcumin, which improved the performance of curcumin on Shigella dysenteriae and reduced the minimum growth inhibitory concentration compared to Kareem's research to 225 μg/mL. Therefore, the nanoization of curcumin particles is more effective than curcumin.
The genes studied in the research can be considered prominent in the pathogenicity of this bacterium. By comparing the results of previous studies with the present study, it is concluded that nanotechnology improves the performance of curcumin particles and creates higher sensitivity in bacterial cells because curcumin is less used due to its poor solubility in water, rapid decomposition and bioavailability. This study investigated the aminoglycoside molecular resistance genes aadE, aacA-aphD, aph Shigella dysenteriae isolated from children and their expression under the influence of curcumin nanoparticles. Based on the results of this study, curcumin nanoparticles at a concentration of 225 μg / mL inhibit the growth of Shigella dysenteriae bacteria. The Sub MIC concentration of curcumin nanoparticles (128 μg/mL) was used to investigate the expression of the aadE gene. By comparing isolates containing aadE gene treated with curcumin with untreated isolates, the expression of this gene decreased, and the relative amount of Fold change was recorded in treated isolates of -1.03. In general, it can be concluded that using an appropriate dose of curcumin can effectively reduce the expression of aminoglycoside resistance genes and be used in clinical cases.
The authors of the article express their gratitude to the staff of Pasargad Microbiology Research Laboratory (Tehran), especially Dr. Amini, and all the loved ones who had useful and valuable guidance in carrying out this scientific activity.
This article is extracted from a master's thesis with research code 1212900690859001400162493751 registered in the Islamic Azad University of Arak. This article has the code of ethics with the number IR.IAU.ARAK.REC.1401.003.
Conflicts of Interest
The authors declare any conflict of interest.
All authors contributed to data collection, writing, and manuscript revision. All authors read and approved the final manuscript.
None.
Received: 2022/06/7 | Accepted: 2022/08/12 | ePublished: 2023/01/20
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |








