BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1184-en.html
2- Department of Biology, Sanandaj Branch, Islamic Azad University, Sanandaj, Iran , f.keshavarzi@iausdj.ac.ir
3- Department of Medical Microbiology, Faculty of Medicine, Hamadan University of Medical Sciences, Hamadan, Iran
Introduction
Gastric Ulcer is a painful injury in the internal gastric wall or the initial part of the small intestine. There is no clear reason for the gastric ulcers. Today, it is evident that gastric ulcer is due to an imbalance of gastric and duodenal ulcers. The main bacterium involved in gastric ulcers is Helicobacter Pylori (H. pylori) (1-4).
H. pylori is a gram-negative and spiral-shaped bacillus. The size of this bacterium is around 0.3  5.5 micrometers. These bacteria are colonized in the human stomach epithelium and most of the other animal species. They have an important role in pathogenicity of gastric inflammation, gastric ulcer, and gastric cancer. They are also etiologically important in the illnesses of peptic ulcer and malignant stomach (5-7).
5.5 micrometers. These bacteria are colonized in the human stomach epithelium and most of the other animal species. They have an important role in pathogenicity of gastric inflammation, gastric ulcer, and gastric cancer. They are also etiologically important in the illnesses of peptic ulcer and malignant stomach (5-7).
Antrum mucus, exotic, and cardia are equally sensitive to infection and the bacteria are colonized in the same way (8). The bacteria of H. pylori are colonized in the stomach of half of the population in the world. Tissue gastritis is a public complication among all individuals infected with these bacteria; however, it has evident clinical circumstances similar to gastric ulcer and gastric cancer just among a few of the patients (5-9).
The mechanisms responsible for different stages of the disease are not fully understood. However, two pathogenicity indices have been identified which have been introduced as virulence factors of different bacterial strains (10-11). These factors are cytotoxin-associated gene A (cagA) and vacuolating cytotoxin (vacA) (12-14).
The cagA gene is located in the region of I in the pathogenicity part (CagA, PAI CagA) and is one of the leading causes of pathogenicity. Cag PAI is a 40 kb locus and its percentage of guanine-cytosine (35% to 39%) is different from the other genomes (8) and is transmitted through some strains of H. pylori. This island has 31 genes out of which, 6 are coding the fourth type of secretion system and take the role in the association of the host bacteria with the pathogenicity process (1-10).
CagA is a 120–145 kDa protein encoded on the 40 kb cag pathogenicity island (PAI). The cagA protein is one of the superficial proteins of the external membrane in H. pylori that has great power in raising the safety. The products of this gene cause duckling in the host cell, after entering bacteria to the stomach epithelial cells. The cagA protein can increase the virulence of bacteria after entering the target cell directly and interference with the cell messaging systems. The existence of this gene in some strains of H. pylori shows the existence of such pathogenicity island of cag PAI (6-9).
H. pylori strains can be divided into cagA-positive or -negative strains. It seems that the severity of digestive illnesses such as peptic ulcer, gastric mucosal atrophy, and gastric cancer is because of H. pylori relating to cag pathogenicity island such as peptic ulcer (1-15). Also, studying the specific bacterial genes of glmM was used to prove, control, and study the existence of H. pylori. According to the previous studies, the glmM gene had more sensitivity to recognize these bacteria in comparison with the other genes (10). Therefore, the purpose of the present survey was to study the frequency of the cagA gene of H. pylori gene among the patients suffering from gastric ulcers in the City of Kermanshah.
Materials and Methods
Samples
The present research was a case-observing study. The samples were 75 PA paraffin blocks; 40 men (47±28) and 35 women (47±26) of the patients suffering from gastric ulcer that were randomly collected from Imam Reza Hospital in the City of Kermanshah, Iran. Participants in research clearly expressed consent to being involved in the study. Then, the information related to each patient was collected from the registered Office of Pathology in the Hospital.
Selecting the patients was performed randomly, and the only inclusion criterion was gastric ulcer.
DNA Extraction
DNA extraction was done by the boiling method according to the Protocol. Then, genomic DNA samples (4 µL) mixed with diluted loading dye (2 µL) were electrophoresed on 1.5% agarose gel.
Measuring DNA Concentration
The optical absorption of extracted DNA samples was measured by NanoDrop (2000/2000C, Thermo, USA) using a ratio of OD260/OD280 to ensure the desired quality of DNA to be used in PCR.
Molecular Studies
The primers sequences to amplify the glmM gene were selected from a previous study (16) and primers for the cagA gene were designed by Gene Runner software considering all the points of SNP. To ensure the accuracy of the primers, they were blasted in NCBI database (Table 1). Primers were synthesized by Cinnagen Company (Iran). Molecular study was performed by PCR method using the PCR detection Kit (Cinnaclon). The PCR cycles of the desired genes are presented in Table 2, separately.
Table 1. The primers sequences
| Length of the piece | Primer connection temperature | Primer sequence | Primer name |
| 294bp | 58 | AAGCTTTTAGGGGTGTTAGGGGTTT AAGCTTACTTTCTAACACTAACGC |
glmM-F glmM-R |
| 183bp | 55 | TTGACCAACAACCACAAACCGAAGC TTCCCTTAATTGCGAGATTCC |
CagA-F CagA-R |
Table 2. PCR proliferation conditions
| Cycling condition | |||||
|---|---|---|---|---|---|
| Final Extension | Extension | Annealing | Denaturation | Initial denaturation | Genes |
| 72 °C 5 min |
72 °C 30Sec |
58 °C 60 Sec |
94 °C 30 Sec |
94 °C 5 Min |
glmM |
| Repeated for 35 Cycles | |||||
| 72 °C 5 min |
72 °C 30Sec |
55 °C 30 Sec |
94 °C 30 Sec |
94 °C 5 Min |
cagA |
| Repeated for 35 Cycles | |||||
Statistical Analysis
The data were analyzed by Excel software and SPSS 20 (SPSS Inc., Chicago, Ill., USA) and frequency of the genes were studied. Also, the logistic regression test was used to determine the association of each form of the disease with the virulence genes alone. A P-value<0.05 was considered statistically significant.
Results
The electrophoretic patterns of The PCR products of glmM and cagA genes were presented in Figures 1 and 2, respectively. The length of amplified fragments related to glmM and cagA genes are 294 and 183 bp, respectively. After confirming glmM-positive samples, which indicate the presence of H. pylori, these samples were used to monitor and determine the presence of cagA gene.
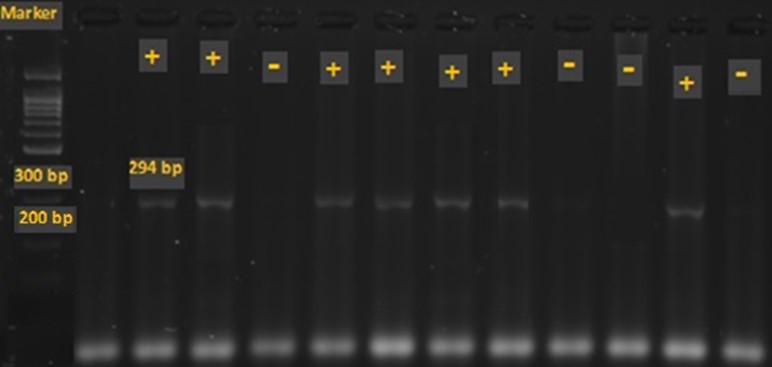
Figure 1. The electrophoretic pattern of the PCR product of glmM gene. The length of amplified fragment by glmM primers is 294 bp. The first column shows the ladder of 100 bp. The samples of 2, 3 and 5-8 and 11 are positive for glmM and 1, 4 and 9, 10, 12 are negative for this gene.

Figure 2. The electrophoretic pattern of the PCR product of the cagA gene. The length of amplified fragment by cagA primers is 183 bp. The first column shows the ladder of 100 bp. The samples of 1,2,3 4 and 7,8,9 are positive for cagA and 5 and 6 are negative for this gene.
The results showed that out of 75 samples, 56 were positive for glmM gene and out of 56 positive samples (glmM-positive), 39 (69.64 %) were positive for cagA gene.
Among them, the frequency of glmM gene was more in men (29, 38.66%) compared to women (27, 36%), although not significant. Also, regarding cagA gene, its frequency was more in men (22, 39.28%) compared to women (17, 30.35%) but not significant (Table 3) (Figure 3).
The correlation between the clinical forms of the disease and the presence of glmM and cagA genes was found to be significant (P=0.005).
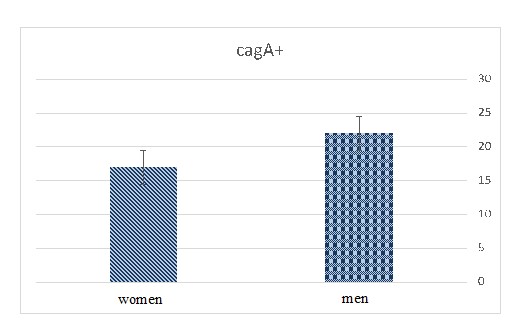
Figure 3. Comparing the frequency of positive cagA gene
Table 3. Comparing the frequency of positive glmM and CagA genes
| GlmM+ | GlmM-- | CagA+ | CagA- | |
| Patients N=75 |
56 (74.66%) | 19 (25.33%) | 39 (69.64%) | 17 (30.35%) |
| Man 40 (53.33%) |
29 (38.66%) | 11 (14%) | 22 (39.28%) | 7 (12.5%) |
| Woman 35 (46.66%) |
27 (36%) | 8 (10.6%) | 17 (30.35%) | 10 (17.85%) |
The study of glmM gene among women and men indicated that 23 patients (30.66%) under 40 years old, and 33 patients (44%) over 40 years old tested positive for glmM gene that shows the frequency of this gene is more among patients who were over 40 years old. Also, about cagA gene, 16 patients (21.33%) under 40 years old and 23 patients (41.07%) over 40 years old were positive for the cagA gene that shows the frequency of this gene is higher in the patients over 40 years old (Table 4).
Also, the relation between age and sex of the patients with clinical forms of H. pylori disease was studied using Chi-square and Fisher's exact tests. No significant association was found in this regard.
Table 4. Comparing the frequency of the positive glmM and cagA genes among people with the age around 40 years old.
| Patients | GlmM+ | GlmM-- | cagA+ | cagA- |
| N=75 | 56 (74.66%) | 19 (25.34%) | 39 (69.64%) | 17 (30.35%) |
| Less than 40 | 23 (30.66%) | 11 (14.64 %) | 16 (28.57 %) | 7 (12.5 %) |
| More than 40 | 33 (44 %) | 8 (10.70 %) | 23 (41.07 %) | 10 (17.85 %) |
Discussion
In this research, 75 paraffin blocks of stomach biopsy samples from the patients infected with H. pylori and suffering from gastric ulcers were investigated in the province of Kermanshah, Iran. Also, the extra information about these patients was achieved through registered Office of Pathology, Imam Reza Hospital.
The glmM gene, which has more sensitivity for H. pylori detection compared to the other genes, was used to identify positive H. pylori samples (10-11). Out of 75 samples, 56 (74.66%) were positive for the glmM gene that indicates the high frequency of H. pylori among patients suffering from gastric ulcer that was in line with the previous studies outcome (8).
In the study of Megrow et al., the prevalence of H. pylori or the prevalence of glmM gene in the biopsy specimens was 44% (12). In glmM-positive samples, the prevalence of cagA was 69.64% (39), indicating a high frequency of this gene in these samples. In previous study by Jafari et al., on 280 gastric biopsy samples in Tehran, the frequency of the cagA gene was about 74.3% (13).
The prevalence of the cagA gene in the studies of Douraqi and Crashia were 84.02% (2) and 41.5% (14), respectively. These results were different from the previous studies in Europe (66-73%) but identical with the majority of studies in Middle East countries (Turkey, Egypt, Israel, and Jordan) in which the outbreak of the cagA gene was between 26 to 44% (7).
In another study by Hussein et al., the frequency of the cagA gene was 65.9% along with achieved results from gathered samples over the same gene in Tunisia (61.6%), Iran (76%), and Iraq (71%) (15), and was similar to the results of the present study. In the study by Essawi et al., which was conducted to recognize the existence of H. pylori in biopsy samples through PCR, the frequency of this gene was 65.9% (16) that accords with my results.
Then, the patients were studied and compared based on the gender. The findings showed the frequency of cagA gene in man at 22 (39.28%) and in women at 17 (30.35%). The frequency of the cagA gene was more in men compared to women. It seems that high number of men samples or having more exposure of men to infection or even the gender and some other factors might be effective.
The patients were also compared in terms of their age. The results showed that the level of cagA gene frequency in the patients under 40 was 16 (21.33%) and in the patients over 40, was 23 (41.07%). This determines that the frequency level of this gene among patients over 40 years old is higher. Thus, the frequency of cagA gene is higher among people over 40 years because of the existence of these bacteria among the elder patients.
Conclusion
In the present study, the frequency of the cagA and glmM genes was investigated in the patients with gastric ulcer in the province of Kermanshah. The results showed high level of glmM gene in the studied patients. Therefore, H. pylori infection is probably an effective factor to cause gastric ulcer in these patients. Also, the high abundance of cagA gene emphasizes the pathogenicity of this gene in positive H. pylori patients. Therefore, monitoring this gene is important in assessing the possibility of gastric ulcer disease.
Acknowledgements
The authors would like to thank the personnel of Imam Reza hospital in Kermanshah.
Conflicts of Interest
The authors declared that they have no competing interests.
Received: 2020/07/14 | Accepted: 2021/05/3 | ePublished: 2021/06/28
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |








