BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1148-en.html

 , Azam Mokhtari
, Azam Mokhtari 
 2, Mohammadreza Mahzounieh1
2, Mohammadreza Mahzounieh1 
 , Somayeh Shahrokh Shahraki1
, Somayeh Shahrokh Shahraki1 
 , Somayeh Gheisarbegi1
, Somayeh Gheisarbegi1 

2- Department of Pathobiology, Faculty of Veterinary medicine, Shahrekord University, Shahrekord, Iran , a.mokhtari@alumni.ut.ac.ir
.
Escherichia coli O157: H7 (E. coli) is an important dietary pathogen that causes dysentery and sometimes hemolytic-uremic syndrome (HUS) (1). This human-animal common human pathogen can remain in food and environments and produce biofilms. Consumption of contaminated foods such as beef, dairy products, ready-made salad, vegetables, and fruits are some of the ways this pathogen is transmitted (3-7). Cow is the primary reservoir of O157 and is a seemingly healthy carrier of this bacterium in the gastrointestinal tract. Cow excretes O157 from its feces, which is the most important risk factor for the contamination of the carcass of this animal (6, 8). Bacteriophage is a type of virus hosted by bacteria. Twort in 1915 and d’Herelle in 1917 discovered bacteriophage and so it has been an option used to control and eliminate bacteria ever since (9). Due to public health concerns about the emergence and increase in antibiotic-resistant bacteria, the use of bacteriophages in the food production and processing industry and medicine has been very welcomed (9-12). Given the high cost of the pharmaceutical industry from discovering new antibiotics, it is essential to develop an alternative treatment regimen that is easy, inexpensive, affordable, robust, and with few side effects to reduce infectious diseases (13, 14).
Studies have shown that phage therapy is successful in the biological control of pathogenic strains of E. coli (18-15) and is superior to other methods because of reproducibility. Unlike antibiotics, when the number of pathogens increases, their specificity, and function can improve. Admittedly, the disadvantage of phage therapy for controlling intracellular endotoxin-producing bacteria is that we may experience an excessive secretion of toxins after the bacteria have died and their walls have been ruptured (17).
Bacteriophage is a new and suitable option to reduce the level of E. coli O157: H7. A review of previous works has shown that E. coli O157: H7 levels decreased in tomatoes, spinach, beef, and meat surfaces after phage therapy (1, 19). Another study has shown that bacteriophage has a reducing effect on the amount of E. coli O157: H7 in cooked and raw beef (20, 21). It is noteworthy that the results of another study identified podophage CA933P as a suitable solution for the removal of enterohemorrhagic E. coli (22).
Therefore, in the current research, we tried to isolate of E. coli O157:H7 bacteriophages from sewage. The results, presenting the isolated phage, can be utilized in developing biocontrol agents against E. coli contamination.
E. coli O157:H7 (ATCC: 35218) prepared from microbial collection of Pasteur Institute of Iran and kept in the microbiology laboratory. It was confirmed as E. coli O157:H7 by PCR assay for stx2 O157:H7 detection. Genomic confirmation was performed using the primers listed in Table 1(23).
Table 1. Specifications of primers used for genomic approval of Escherichia coli (ATCC: 35218)
| Target gene | Primer sequence | Amplicon Size | Reference |
| stx2 | Forward: TTA ACC ACA CCC CAC CGG GCA GT Reverse: GGA TAT TCT CCC CAC TCT GAC ACC |
524 | Pollard, Johnson, Tyler, and Rozee (1990) (24) |
Sampling and Preparation of Sewage
The sampling was conducted via a sterile glass bottle; and sewage samples were taken from a refinery in Chaharmahal and Bakhtiari province. Then the samples were transferred to the laboratory and centrifuged at 8000g for 10 minutes. Finally the supernatant was filtered by sterile 0.2 μm Minisart filters (Sigma- Aldrich, Cat. No.: 16534K).
Preparation of Bacteria and Adding Sewage
At first, one milliliter of overnight bacterial culture medium was added to 20 milliliters of BHI liquid culture medium, and the suspension was incubated for 3 hours at 37°C. Then, 20 milliliters of filtered sewage were added to this suspension and incubated at 37°C for 24 hours. Afterwards, it was centrifuged for 10 minutes at 8000 g. Finally, the supernatant was filtered through a 0.45 μm sterile syringe filter (Sigma- Aldrich, Cat. No.: CLS431225) (24).
Phage Isolation
Bacteria were grown in 20 mL of BHI medium and incubated for 4h at 37°C. Then, 20 mL of the filtered supernatant of the sewage culture in BHI whose preparation steps were added to the bacteria culture and incubated for 24 h at 37°C. After Centrifuge for 10 minutes at 8000 g, the supernatant were filtered through a 0.2 μm syringe filters (Sigma- Aldrich, Cat. No.: CLS431229). Phage isolation was performed using the double agar method. First, 9 mL semi-solid BHI (containing 0.7% agar) was placed into tubes and sterilized. When the temperature of the semisolid medium reached to about 45°C, 0.1 mL of overnight cultured and filtered bacteria was added to it and spread throughout the culture medium. This culture medium was then added to a solid and sterile BHI medium (containing 1.5% agar) to form a two-layer culture. When the agar was tightened, 20 μL of the filtered sewage was placed in the center of the culture medium and incubated for 24 hours at 37°C (24).
Electron Microscopy
The phage suspension was centrifuged for 90 minutes at 20000g. The supernatant was then slowly withdrawn from the tube and the pellet adhered to the tube wall was dissolved in 50 mL of Phage buffer and again centrifuged as described above. After centrifuge, the supernatant solution was removed and the pellet was dissolved in 25 mL of Phage buffer. For the coloring of the phages, 10 μL of the suspension was transferred to a carbon-treated copper grid (400 Mesh) and placed for 210 seconds in this mode. Then, the grid was then placed in the room temperature for 20 seconds. After that, 20 μL of uranium acetate was poured onto the grid and after 160 seconds. Eventually, the excess uranium acetate was removed gently using a drying paper, and the grid was left in room temperature for 30 minutes to be completely dried (25).
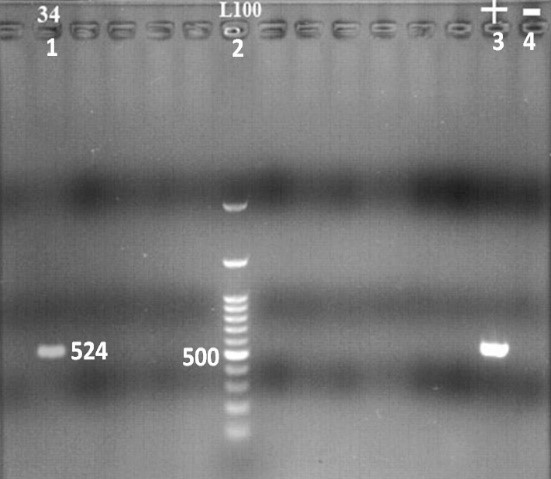
In the current study, for the confirmation of O157:H7, the presence of stx2 fragment of O157:H7 was detected using PCR test. Positive PCR product and positive control sample were in the size of 524 bp, while no bands were detected for negative control after electrophoresis in 1% agarose gel (Figure 1).
Figure 1. Electrophoresis gel image of E. coli O157: H7 PCR product
1: positive sample for stx2 (524 bp), 2: 100 bp leader, 3: positive control (524 bp), 4: Negative control
To detect the plaque formation, lytic bacteriophages were isolated after the inoculation of the sewage into double layer agar, and the phage plaques were completely formed in the plates. These plaques indicated that these phages had lytic effects on E. coli O157: H7 (Figure 2).
Figure 2. Phage plaques on culture medium
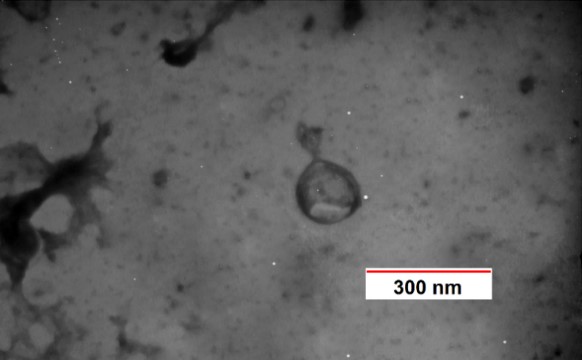
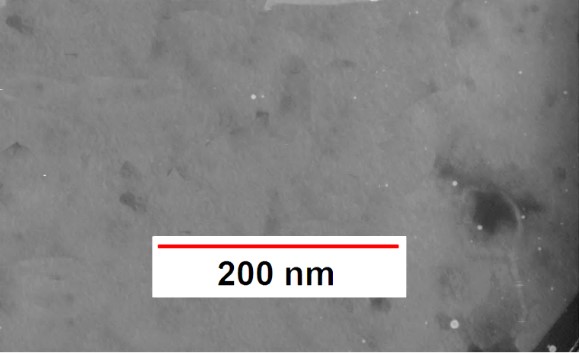
In the images taken by TEM electron microscopy, due to the morphological features, the isolated phages belonged to the Podoviridae, Myoviridae, and Cystiviridae families (Figures 3-5). The podoviridae have symmetrical heads and non-retractable tails. Members of the myoviride family have relatively high symmetrical head and tail contraction, and cystoviride members have spherical heads and contraction tails.
Figure 3. Electronic microscope image of phage belonging to the Cystoviridae
Figure 4. Electronic microscope image of phage belonging to the Podovirida
Figure 5. Electronic microscope image of phage belonging to the Myoviridae
Therefore, in the current study, using TEM microscopy observation, phages against E. coli O157:H7 belonging to the Podoviridae, Myoviridae and Cystiviridae families isolated from the liquid sewage samples were taken from a refinery in Chaharmahal and Bakhtiari province, Iran. The basis of the diagnosis in the present study was the shape and symmetry of the phages, which, of course, must be confirmed by other molecular methods. In the previous studies, E. coli phages have been isolated from different sources. For example, Jurczak-Kurek et al. (2016) detected 60 infecting E. coli bacteriophages from urban sewage. They found the phages belonging to Siphoviridae and Podoviridae families, using virion and plaque morphology evaluation, propagation temperature range and thermal inactivation conditions and the effects of the osmotic shock, high and low pH and detergent or organic solvents, and finally genomic analyses (25). In the present study, due to lack of budget, we only performed virion and plaque morphology evaluation that should be confirmed with more tests.
In another study, Askora et al. (2015) identified an effective lytic bacteriophage against a number of enthraemorrhagic and enteropathogenic Escherichia coli strains. They named the isolated phage as øZE1. It was confirmed that the phage belonged to family Siphoviridae using electron microscopy and produced lysis on four E. coli strains. Also, the resistance to pH, heat, and chloroform was evaluated, and genomic analysis of ØZE1 phage was performed (26). In the present study, similar to the study of Askora et al., the initial basis for the diagnosis of phage was based on the electron microscopy.
Jamal et al. (2015) isolated and characterized a phage belonging to Myoviridae that was effective on antibiotic resistant Escherichia coli strains. In their study, the phage morphology was identified using TE microscopy the same as what we performed in the current research. Furthermore, they described high levels of resistance of MJ1 to heat and pH change (27).
The sufficient therapeutic effect of phages has been established in many studies. For example, Periasamy and Sundaram (2013) applied bacteriophages for pathogen removal from wastewater. In their study, E. coli specific phage was isolated, and its effective titer was standardized. (24). Although, the titration of phages were not performed in the current study, we found antibacterial activity of isolated phages using plaque formation observation.
In another study, Sadekuzzaman et al. (2017) reduced E. coli O157:H7 in biofilms by bacteriophage BPECO 19. The phage treatment performed by Sadekuzzaman et al. decreased E. coli viability (2). Phage treatment, which was used in this study, reduced the survival of O157: h7. With plaque formation test, we found antibacterial activity of isolated phages against E. coli O157:H7.
Arthur et al. (2017) used bacteriophage to reduce the bacterial population in the skin and carcasses of cows found in beef processing plants. The results showed that phage did not significantly reduce E. coli O157: H7 during processing (28). In another work, Seo et al. (2016) inhibited the growth of O157:H7 in beef, pork, and chicken meat by BPECO19 phage (29).
Overall, due to the increasing threat imposed by multidrug resistance, it is necessary to search for novel antimicrobials. There is a growing need for alternative agents of antibiotics and conventional drugs for the prevention and treatment in humans and animals. Exploring bacteriophages as biological control agents can help control antibiotic resistant pathogens. In addition, bacteriophage may apply in pathogen detection and biopreservation. In the present study, E. coli O157: H7 infecting phages were isolated and were available for complementary studies assaying the biological aspects of isolated phages and their in vivo and clinical applications. In addition, further studies can be helpful to use the potential for therapeutic potential and evaluate isolated phages for virulence factors and their ability to transmit genes.
Overall, in the current study, E. coli O157:H7 phages belonging to the Podoviridae, Myoviridae and Cystiviridae families were isolated from liquid sewage samples and are available for the future studies.
This work was supported by grants from Shahrekord University (Grant number: 98GRD30M1801)
Authors declared no conflict of interests.
Received: 2020/05/23 | Accepted: 2020/11/2 | ePublished: 2021/01/10
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |