BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1132-en.html


 , Bahram Nasr Nasr Esfahani1
, Bahram Nasr Nasr Esfahani1 
 , Ali Mohammad Ahadi2
, Ali Mohammad Ahadi2 
 , Saman Shalibeik
, Saman Shalibeik 

 3
3
2- Department of Genetics, Faculty of Science, University of Shahrekord, Shahrekord, Iran
3- Department of Microbiology, Faculty of biological Sciences, Falavarjan Branch, Islamic Azad University, Isfahan, Iran , saman.shalibeik@gmail.com
.
Coronaviruses, first identified in 1960, are major RNA viruses that affect a wide range of domesticated and pedigree animals as well as bats (1). There are rare studies showing bats can be host of many types of corona viruses. its different due to the area and type of bats (2). These animals appear to be the virus's natural reservoir. Coronaviruses generally cause mild respiratory illness in humans with cold-like symptoms (3, 4). But their ability to cause severe respiratory and even fatal diseases has also been proven. For example, SARS epidemic or acute respiratory distress syndrome during 2002-3 and Corona Middle East Respiratory Virus (MERS-Cov) in 2013, these experiences showed that coronaviruses of animal viruses were able to produce mutated and transmissible species. They have it to humans. Which is usually a severe disease (5, 6). In December 2019, many of the data collected from "unknown viral pneumonia" were suggested, initially related to exposure to the Huanan seafood marketplace, Wuhan, China (7). On January 6, 2020, a new corona virus was discovered that could infect humans and was called COVID-19. As of February 7, 2020, there have been 43,103 confirmed cases of COVID-19 pneumonia in 25 countries (8) COVID-19, like other coronavirus pneumonias, can cause Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) (9). On January 9, 2020, the cause of the disease was a novel coronavirus called nCoV-2019. On January 30, 2020, Corona announced the launch of the new virus, the sixth major public health emergency in the world, a threat not only to China but to all countries (10). On February 11, 2020, the World Health Organization (WHO) chose the official name for the novel coronavirus (COVID-19) and International Committee on Taxonomy of Viruses (ICTV) also changed the name of the virus causing the disease from nCoV-2019 to “severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)” on the same day. This name become chosen because the virus is genetically associated with the coronavirus accountable for the SARS outbreak of 2003. Even as associated, the 2 viruses are one of a kind. WHO introduced “COVID-19” because the call of this new sickness, following recommendations previously developed with the world corporation for Animal health (OIE) and the meals and Agriculture corporation of the United countries (FAO) (11-13). On February 20, 2020, for the first time in Iran, the Ministry of Health announced the positive results of two possible cases of coronavirus in Qom. In this regard, identification groups and management of infectious diseases and rapid response teams have been deployed in this city. An hour later, the deaths of both men were confirmed by the Ministry of Health. On March 23, 2010, the number of victims of Coronavirus in Iran reached 4 and the number of victims reached 18. The World Health Organization (WHO) also announced an international emergency in connection with the outbreak of COVID-19 on January 30, calling the current situation a pandemic. The organization last announced the outbreak of the H1N1 virus in 2009 as a pandemic (14).
Virology
Coronaviruses are the biggest group of viruses belonging to the Nidovirales order, which contains Coronaviridae, Arteriviridae, and Roniviridae families. Coronaviridae are single-stranded, enveloped, nonsegmented, positive-sense RNA viruses named and The size of the virus genome is between 26 and 32 kilobases, which is one of the largest RNA viruses. These viruses have two different types of surface proteins and are named after this apparent feature. There are 4 predominant sub-groupings of coronaviruses, referred to as alpha, beta, gamma, and delta. Human coronaviruses have been first identified inside the mid-1960s. The seven coronaviruses which can infect humans are: commonplace human coronaviruses like 229E, NL63 (alpha coronavirus), OC43, HKU1 (beta coronavirus) and other human coronaviruses as MERS-CoV, SARS-CoV, SARS-CoV-2 (beta coronavirus) (17-15). SARS-CoV-2 is the third coronavirus after the two SARS and MERS viruses in the remaining a long time to infect human beings by way of crossing animal species (15,18). In general, alpha and beta-coronaviruses in particular infect mammals and purpose human and animal diseases. Gamma coronaviruses and delta coronaviruses, however, in particular infect birds; but, a few also can infect mammals. In addition to the human coronaviruses mentioned above, alpha coronaviruses include various bat coronaviruses, rodent coronaviruses such as mice, weasel coronavirus and swine epidemic diarrhea virus, and beta coronaviruses include various mouse coronaviruses and multiple bat coronaviruses. (18,19). The coronaviruses genome, which varies in length from approximately 26,000 to 32,000 bases, includes a number of variables (from six to eleven) that open the open reading frame (ORF). The primary ORF, which makes up approximately sixty-seven percent of the total genome, encodes sixteen non-structural proteins (nsps), even as the closing ORFs encode sub-proteins and structural proteins. Coronaviruses are named for the crown-like spikes on their floor. The 4 major structural proteins encompass spike surface glycoprotein (S), coating protein (E), membrane protein (M), and nucleocapsid protein (N). The 8 sub-proteins include 3a, 3b, p6, 7a, 7b, 8b, 9b, and orf14 (Figure 1) (20-23). Spike floor glycoprotein plays a vital function in binding to receptors at the host cell and determines host motion. The SARS-CoV and MERS-CoV spike proteins are linked to one of a kind host receptors thru different domain names linked to the receptor (RBD). SARS-CoV uses the enzyme angiotensin-changing enzyme 2 (ACE2) as One of the major receptors, the usage of CD209L as an opportunity receptor (24), at the same time as MERS-CoV uses dipeptidyl peptidase four (DPP4), in addition to CD26 is known to apply as the main receiver. The SARS-CoV-2 genome has been mentioned to be more than 80% equal to the preceding human coronavirus (SARS-like bat CoV), and in some areas differs substantially from the SARS coronavirus genome (21-24).
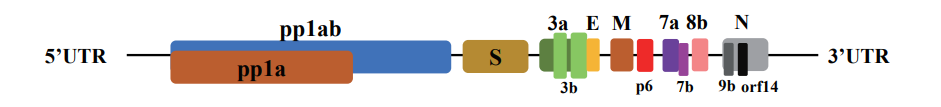
Figure 1. Organizing the SARS-CoV-2 genome: large genes, the ORF1a and ORF1b genes, which encode 16 non-structural proteins (nsp1-nsp16) that are fairly covered in all coronaviruses. Structural genes encode structural proteins, Spike (S), Envelope (E), Membrane (M), and Nucleocapsid (N), and 8 sub-proteins, which includes 3a, 3b, p6, 7a, 7b, 8b, 9b, and orf14. (20)
As determined by genomic research and the presence of some stay bats and animals in the seafood market in Wuhan, the brand SARS-CoV-2 can be derived from bats or bat-inflamed feces in or around the marketplace (25). According to current information, the original and natural host of the new coronavirus appears to be bats, and the virus then enters a medium-sized host (possibly an anteater) (11, 26) or other wildlife. For sale in the market, Wuhan (27) has been transferred from this host to the human being. Then the process of human-to-human transmission begins and the present epidemic is formed (Figure 2). The fact that the anteater is specifically named as the intermediate host is due to the fact that 70% of the anteaters were positive for coronavirus. In addition, the analysis of Whole genome sequencing (WGS) of Pangolin anteater virus showed a nucleotide similarity of 91.02% with the new coronavirus SARS-CoV-2. The sequence of metagenomics sequences of coronaviruses associated with Pangolin scaly anteaters belongs to two subspecies of corona viruses associated with SARS-CoV-2. The discovery of several common lines between the corona of the Pangolin virus and their similarity to the now widespread SARS-CoV-2 in the world suggests that the Pangolins should be considered as possible hosts for the outbreak of the novel corona virus (28).

Figure 2. Origin and transmission of SARS-CoV-2, Possible zoonotic transmission source: media reports and open data
Although primary studies show the relationship between local marketing and wild animals which can probably cause transferring virus from animal to human, other studies show that it transfers mostly from human-to-human by droplets or direct contact. Evidence of a rapid growth in infections and the possibility of transmission via asymptomatic carriers of novel coronavirus may be successfully transmitted to human beings and display high potential for epidemics (29) similarly to the high efficiency of SARS-CoV-2 transmission. The development and comfort of worldwide journey can similarly expand it around the world (29 ,30).
Clinical manifestations
In a study of Guan and et al. Reported 1,099 cases of SARS-CoV-2 infection, with fever and 67.7% coughing in 87.9% of cases, and diarrhea (3.7%) and vomiting (5%). It was rare, accounting for 82.1% of lymphopenia patients and 96% of SARS-CoV-2 infected patients with chest CT images (31). The symptoms of COVID-19 are various, from asymptomatic to acute anxiety syndrome and multifunctional dysfunction. The most commonplace symptoms at onset of COVID-19 consist of fever (not in all cases), headache, cough, fatigue, sore throat, myalgia and shortness of breath. Conjunctivitis is likewise described. Consequently, they may be indistinguishable from different respiratory infections (32). In a subset of patients, via the end of the primary week, the disorder can lead to pneumonia, respiration failure, and loss of life. This development is observed by using a sharp boom in inflammatory cytokines which include IL2, IL7, IL10, GCSF, IP10, MCP1, MIP1A, and TNFα (33). The median time from onset of symptoms to dyspnea was 5 days, hospitalization 7 day and acute respiratory distress syndrome (ARDS) 8 day. Recuperation started at 2 or three weeks. The average hospital remains for people who recovered turned into 10 days. Side effects and mortality are greater common in the aged and people with joint disorder (50-57% of fatalities). Mortality rates in person sufferers ranged from 4-11%. The overall mortality rate is anticipated to be among 2 and 3 % (34). Apparently, the disorder has been suggested to be milder outside Hubei Province than in patients in Wuhan (35). In addition, the mortality has been reported to be milder in patients outside Hubei Province than in patients in Wuhan (36). This can be due to the truth that during the ones instances suggested from Wuhan handiest intense instances or due to the predisposition of the Asian population to the virus because of in addition expression of ACE2 receptors within the breathing mucosa (37). It has also been reported that the disease is milder in infants and children than in adults. In one study, 34 kids admitted to Shenzhen health center in China among January 19 and February 7, together with 14 males and 20 females. The average age of 8 years and 11 months was observed in 28 children who were infected by a family member and in 26 children with a history of travel or residency in Hubei Province, China. All sufferers had been both asymptomatic (9%) or had mild disorder. No intense or extreme cases were reported. The maximum common symptoms have been fever (50%) and cough (38%). All sufferers recovered with treatment symptoms and there have been no deaths. A case of intense pneumonia and dysfunction in a baby has also been said (38). Further, infant reported cases were slight (39).
Relationship between the ABO Blood Group and the COVID-19 Susceptibility
In a study by Jiao Zhao and et al. Of the relationship between ABO blood type and COVID-19 susceptibility, the number of patients and victims of covid-19 who had blood type A was significantly higher than the number of healthy people in the same group. It's bloody. By comparison, fewer people with blood type O had the disease or died. Blood groups compared 2,173 definite patients with COVID-19 in the cities of Wuhan and Shenzhen with 3,694 healthy individuals in the Wuhan region. While 31.16 percent of Wuhan residents had blood type A, 37.75 percent of patients with coronavirus had the same blood type at Winy Hospital in Jinyintan Hospital, compared with only 25.8 percent of patients with 33.84 percent of healthy people with blood type O The hospital had a blood type. The researchers also looked at 206 patients who died of the coronavirus and found that 85 victims, or 41.26 percent, had blood type A. Only 52 of the dead (a quarter) had blood type O. This research does not mean that people with blood type A are 100% infected and that people with blood type O do not have 100% immunity. Further research is needed to draw definitive conclusions (40).
COVID-19 diagnosis
Diagnosis of COVID-19 disease is based on radiological and laboratory results. Radiological studies are important in early diagnosis of COVID-19. The main feature of radiological images in patients with ground glass opacity and lung density is consolidation. CT scans of the chest in patients with COVID-19 usually show ground-glass with or without consolidation, according to viral pneumonia (11). Case studies have shown that chest CT abnormalities are much more likely to be bilateral, have an environmental distribution, and contain the lower lobes. Much less common findings include thickening of the pleura, pleural effusion, and lymphadenopathy. In the United States, the CDC recommends collecting samples for SARS-CoV-2 testing from the upper respiratory tract (nasal-pharyngeal and oral-pharyngeal swabs) and, if possible, the lower respiratory tract (sputum, chip aspiration, or bronchoalveolar lavage). Induction of sputum is not indicated. Additional samples (such as stool, urine) can also be collected. Respiratory sampling should be performed with caution in terms of the risk of airborne transmission. Conventional diagnostic tests, such as assessment for the detection of antiviral antibodies or viral antigens, have been clinically developed and used (41). A diagnostic confirmatory test is usually an RT-PCR that identifies the ribonucleic acid virus genome, and the area identified by this technique includes the RdRp genome region along with the E or N genomic regions (42, 43). In a study of 1,014 patients in Wuhan who underwent both reverse polymerase chain reaction (RT-PCR) and chest CT tests for COVID-9 evaluation, a positive chest CT for COVID-19 (according to consensus) There were two radiologists) using PCR as a reference test, with a sensitivity of 97%, although the specificity was only 25%. Low specificity may be related to other causes of CT-like findings (44).
Treatment Options for Coronavirus Disease 2019 (COVID-19)
As of April 18, 2020, the virus has infected more than 2275783 people worldwide and killed more than 156104 people (45). Up to now, no specific treatment has been powerful for SARS-CoV-2 infection. Supportive care, such as oxygen in mild instances and extracorporeal membrane oxygenation for severely ill sufferers, particular medications for the ailment are nevertheless being taken into consideration. The production of safe and stable vaccines is a major challenge, and research and development of new drugs is a very long process. The key is supportive care, such as maintaining vital signs, regulating oxygen and blood pressure, and reducing complications such as secondary infections or organ failure. In such a sudden epidemic, scientists were unable to produce new drugs in accordance with traditional principles (46, 47). Antiviral drugs, such as FDA-approved drugs such as penciclovir, nitrazine, nafamostat, chloroquine (chloroquine), and two broad-spectrum antivirals and remdesivir antiviral drugs with effects on cytotoxicity, virus function, and SARS-CoV-2 infection rates have been evaluated (47). Chloroquine has been well described with in vitro results in uncoating inhibition and put up-translational change in newly synthesized proteins, specially glycosylation inhibition in lots of viruses, including immunodeficiency virus. Recent results have shown that Remdesivir and Chloroquine are effective in controlling SARS-CoV-2 infection in laboratory conditions and can be evaluated in Covid-19 disease. Similarly, to 1 case of Covid-19 pneumonia with promising clinical responses to Remdesivir and clinical trials in China, more medical research than Remdesivir are needed to confirm its healing efficacy (48, 49). Remdesivir is currently used to treat Ebola virus infection. It is in the process of clinical research. Holshue et al. Used Remdesivir within the treatment of sufferers with SARS-CoV-2 infection and carried out good consequences (50) In the United States, the first patient infected with SARS-CoV-2 was treated before recovery and rehabilitation through supportive care and intravenous Remdesivir. Teicoplanin is a glycopeptide antibiotic commonly used to treat bacterial infections and has been shown to be effective against the virus in the laboratory. The antibiotic has recently been used to treat gram-positive bacterial infections, especially staphylococcal infections, and has previously been shown to have antiviral effects on Ebola, influenza and the hepatitis C virus, and on coronaviruses such as MERS and SARS. A treatment model has been developed to treat MERS infection. Based on a study of corona viruses by Baron et al., Teicoplanin acted on the early stages of the virus's life cycle by inhibiting the low-pH cleavage of the virus's cuneiform protein using catapsin L in the endosomes and releasing genomic viral RNA. And it prevents the virus from multiplying. It has recently been used to treat gram-positive bacterial infections, especially Staphylococcal infections and It has previously been shown to have antiviral effects on Ebola, influenza and the flavivirus of hepatitis C viruses (HCV) and on coronaviruses such as MERS and SARS and further study of the antiviral effects of this molecule on the causative agent of COVID-19 is recommended (51).
In a research done by Chang Chen et al. to examine the effect of Favipiravir (FPV) against Arbidol in the treatment of COVID-19. The results of their study showed that FPV was more effective in reducing fever than Arbidol. FPV is an RNA-dependent RNA polymerase (RdRp) inhibitor that has been shown to be effective in treating influenza and the Ebola virus. Favipiravir has not yet been included in the COVID 19 treatment protocol (52). The Fifth Edition of Infection Prevention and Control (IPC) states that patients with severe and critical illness can be treated with Convalescent plasma (plasma with antibodies from recovered COVID-19 patients) (53). hrsACE2 A new drug has been discovered by a group of scientists around the world and has entered the second phase, raising hopes for a cure. Angiotensin-converting enzyme (ACE2) enzyme 2 is an important receptor for SARS-CoV. ACE2 is now known to be the main recipient of SARS-CoV-2 infections, and it has been suggested that inhibition of this interaction may be used in the treatment of patients with COVID-19. In this study, it was shown that SARS-CoV-2 can be propagated directly in the blood vessels and kidneys in addition to the lungs. Laboratory cell culture results have shown that hrsACE2 can reduce the amount of virus in infected tissue by between 1,000 and 5,000 times that of normal conditions. The results of these studies showed that SARS-CoV-2 could directly engineer human blood vessel organoids and infect human kidneys, which could be inhibited by hrsACE2. These data suggest that hrsACE2 can significantly block the early stages of SARS-CoV-2 infection (54). However, the best way to deal with the epidemic is now to control the sources of infection.
The outbreak of COVID-19 has come to be a medical threat to the overall population and health care personnel round the world. However, knowledge approximately this novel coronavirus is restricted. The effective antiviral remedy and vaccination choice is currently being evaluated and developed. What we can do now could be aggressively put into effect infection control measures to prevent the unfold of current coronavirus via human-to-human transmission. The greater information to be had about the novel coronavirus and its occurrence, the better it'll be able to cope. It is hoped that with the discovery of the vaccine, drugs and therapeutic measures affecting SARS-CoV-2 will be overcome in the near future.
Suggestions: There is still a lot to be learned about how COVID-19 spreads, its severity, and other features of the virus; epidemiological and clinical research is underway and the situation is changing rapidly. However, due to the incubation period of the virus and its transmission from one person to another, the question arises as to whether it will lead to virus attenuation. Epidemiological models show that interventions to isolate patients and "social distancing" in order to reduce contact with individuals in society greatly affect the number of new cases of covid 19. Due to the lack of vaccines and appropriate treatment, the best way to prevent illness is to avoid being exposed to this virus. The virus is thought to spread mainly from human-to-human through respiratory droplets produced when an infected person coughs, sneezes or talks.
The authors thank all those who helped them writing this paper.
Authors declared no conflict of interests.
Received: 2020/03/5 | Accepted: 2020/04/15 | ePublished: 2020/05/20
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |





