BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
URL: http://ijmm.ir/article-1-1110-en.html
2- Department of biology, Islamic Azad University, Ardabil Branch, Ardabil, Iran , royasafarkar@yahoo.com
3- Department of biology, Islamic Azad University, Guilan Branch, Guilan, Iran
.
Acinetobacter baumannii is an opportunistic pathogen that causes a wide range of infections including pneumonia, bacteremia, urinary tract infection, wound infection, and meningitis (1). A.baumannii has the ability to survive in the hospital environment and resist against various antibiotics, which causes a high prevalence of nosocomial infections (2). The ability to form biofilm on medical equipment and devices is a significant agent in the pathogen of this bacterium (3). Many factors such as outer membrane protein A (OmpA), biofilm-related protein (Bap), beta-lactamase PER-1, etc. are involved in the formation of biofilm (4). Some of surface proteins like ompA, blaPER-1, and Bap in addition to being involved in biofilm formation, are also involved in bacterial binding to human epithelial cells and non-living surfaces (5(. Biofilm is a complex association of microbial colonies that lead to the formation of a cellular matrix, which is formed of a protective layer of polysaccharides (6). The sticky cell in the biofilm is located within an extracellular matrix, which is containing extracellular polymeric materials (EPS). Microbial cells are the main parts of biofilm that produce EPS, which include 50 to 90% of the total organic carbon in biofilm (7). So, biofilm formation is the main agent in bacterial survival and resistance (8). The bap gene is one of the most important biofilm producing and amplifying genes in A. baumannii. This protein with high molecular weight is located on the outer surface of the bacteria and contains a central nucleus including the successive repeats of similar sequences (9, 5).
There are various methods to evaluate the ability of biofilm formation by A. baumannii, which include the Agar Congo Red, Tube method, and Microtiter plate quantitative method that is the gold standard method for biofilm production (10). Molecular methods such as PCR have recently been applied to investigate and identify the genetic factors in biofilm formation. A. baumannii by expressing the bap gene decreases permeability and thereby causing resistance to a variety of bacteria, also causes high stability of the pathogen by creating high adhesion to biotic and abiotic surfaces (11).
The increasing prevalence of antibiotic-resistant isolates of A. baumannii producing biofilm in recent years in different geographical areas has led to failure in the treatment process. Considering the high prevalence of nosocomial infections caused by A. baumannii and also the spread of factors that increase resistance, the aim of this study was to investigate the frequency of bap and blaOXA-51 genes in A. baumannii isolates, which isolated from nosocomial infections in Tehran.
Collection and identification of bacterial isolates
In this descriptive cross-sectional study, after approval in the ethics committee with the code IR.ARUMS.REC.1398.024, from May to November 2019, 165 isolates from blood, urine, trachea, sputum, pus, and wounds of hospitalized patients in different hospitals of Tehran were collected with 95% confidence level and 5% error rate.
Each isolate was cultured on McConkey agar and blood agar in the laboratory and was incubated for 24 hours at 37°C. Next, the presence of Gram-negative coccobacilli of Acinetobacter was confirmed microscopic direct Gram stain test. In order to detect the different species of Acinetobacter, IMVIC, catalase and oxidase, SIM, VP, OF, TSI, urease and the growth was carried out at 37 and 42°C. The isolates with the reaction of lactose-negative, immobile, oxidase-negative, catalase-positive, indole-negative, pigment-negative, urease-negative, citrate-positive, H2S-negative, and VP-negative were isolated as A. baumannii. Furthermore, the blaOXA-51 gene which is intrinsic to A. baumannii for final confirmation of the isolates in all isolates was detected.
Phenotypic antibiotic susceptibility test
Determination of antibiotic susceptibility of the collected isolates was done by the disk diffusion method using antibiotic discs made by the company (Padtanteb, Iran) on Muller Hinton agar (Merck, Germany) and according to CLSI 2019 instructions (12). Tested antibiotics include: tobramycin (10 micrograms), cefepime (30 micrograms), imipenem (10 micrograms), meropenem (10 micrograms), amikacin (30 micrograms), gentamicin (10 micrograms), ciprofloxacin (5 micrograms), Were cefotaxime (30 μg) and ceftazidime (30 μg). In this method, a bacterial suspension with turbidity equivalent to a half McFarland was inoculated on the Muller-Hinton Agar medium in three directions. The cultured plates were kept at laboratory temperature for 20 minutes to remove excess moisture in the culture medium. Then, after placing the discs on the culture medium and incubating for 24 h at 37°C, the diameter of the growth inhibition zone for each antibiotic was recorded as sensitive, semi-sensitive and resistant according to the relevant instructions.
Genotypic evaluation of the presence of blaOXA-51 and bap genes
DNA extraction
To extract DNA, one to three colonies of bacterial isolates were boiled in 100 μL sterile water for 15 minutes and then placed in a bowl of ice for 15 minutes to induce a cold shock. Finally, it was centrifuged at 11,000 rpm for 2 minutes (13). After extraction, the DNA samples were kept at -20°C until doing PCR. Determining the Quantity of DNA extracted by nanodrop device (Thermo-2000c) was investigated.
Polymerase chain reaction (PCR)
The Polymerase chain reaction was used to investigate the presence of blaOXA-51 and bap genes. To achieve this, oligonucleotide sequences of specific primers for these genes were used (14), which is shown in Table 1. In this study, a reaction mixture (Master mix_ CinnaGen) was used which included 1 μL of each primer (CinnaGen), 13 μL of the reaction mixture, 8 μL of double distilled water and 1 μL of DNA template were added and the final volume of the compounds was increased to 25 μL. And the mixture was placed in a thermocycler device (BIOER_Model: TC-XP-G). The program process of PCR cycles of blaOXA-51 gene was as follows: primary degeneration for 5 minutes at 95°C and then 30 thermal cycles, including degeneration for 45 seconds at 94°C, binding of primer to DNA template for 45 seconds at 55°C, template strand elongation for 60 seconds at 72°C and final elongation temperature for 10 minutes at 72°C. The bap gene was replicated corresponding to the blaOXA-51 gene, but at 1.45 seconds to elongate template strand.
Electrophoresis of PCR products was adjusted by 1.5% gel containing 1% DNA safe stain (CinnaGen) at 120 volts for 45 minutes and after this time the bands were photographed by transilluminator gel (E-BOX_VX5). A. baumannii ATCC 19606 and a microtube containing reaction materials without the DNA template were applied as positive and negative controls, respectively.
| Source | Size(bp) | Primer sequence | Primer |
| (14) | 350 | F:5′-TAATGCTTTGATCGGCCTTG-3′ R:5′- TGGATTGCACTTCATCTTGG-3′ |
blaOXA-51 |
| (14) | 1449 | F:5′- ATGCCTGAGATACAAATTAT-3′ R:5′- GTCAATCGTAAAGGTAACG-3′ |
bap |
Phenotypic Investigation of Biofilm Production in A. baumannii Isolates
To investigate the biofilm formation ability of the strains, the tubular qualitative (TM) plate method and the quantitative tissue culture (TCP) method were applied.
Phenotypic Evaluation of biofilm production by microtiter plate method
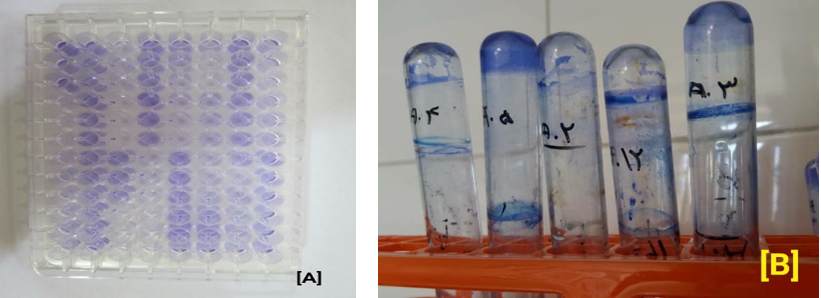
A bacterial suspension equivalent to 0.5 McFarland was prepared in a nutrient broth medium and 200 μL was poured into each well. The microtiter plates were heated for 24 h at 37°C. After the heating period, the contents of each well were aspirated and the wells were washed 5 times with sterile phosphate-buffered saline to remove all planktonic cells. After the wells were dried, 200 μL of 2% crystal violet was added to the wells and after 5 minutes, the wells were aspirated and washed with water. After the wells dried, 160 μL of acetic acid was added to bleach into the wells.
Finally, it was read by an ELISA plate reader at 650 nm. Then, according to Table 2, optical density (OD) of the isolates was compared to control optical absorption and reported as strong, medium, weak, and negative biofilm (15). Pseudomonas aeruginosa PA01 was used as a positive control in biofilm formation tests.
| Lack of biofilm formation | OD ≤ OD (c) |
| Weak biofilm | OD (c) < OD ≤ 2 × OD (c) |
| Moderate biofilm | 2 × OD (c) < OD ≤ 4 × OD (c) |
| Strong biofilm | 4 × OD (c) < OD |
Phenotypic Evaluation of Tubular Biofilm Production
First, 5 mL of the nutrient broth medium was transferred to sterile test tubes, and then 0.1 mL of microbial suspension was added to the tubes and heated at 37°C for 24 hours. After heating, the contents of each tube were aspirated and each tube was washed 5 times with phosphate-buffered saline. After drying, 10 mL of 2% crystal violet solution was added to each tube and after 5 minutes, the dye of crystal violet was removed and the tubes were washed and extracted with 33% acetic acid. Biofilm formation was analyzed qualitatively using this method. The intensity of the color remaining on the tube wall illustrates the formation of biofilm (16). Pseudomonas aeruginosa PA01 was used as a positive control in analyzing biofilm formation tests.
Statistical Analysis
Data were analyzed using SPSS software version 22 (SPSS Inc., Chicago, IL., USA) and t-test at a 95% confidence interval (P<0.05). And the Statistical correlation between biofilm formation ability in A. baumannii isolates was determined by the presence of bap, blaOXA-51 genes, and the antibiotic resistance pattern of the isolates.
Results
The characteristics of the isolates according to the type of sample and the patient’s sex are presented in Table 3.
Table 3. Characteristic regarding the number and percentage of the sample distribution and sex of patients among A. baumannii isolates
| Sample isolation place | Number of isolates divided from patients | Number of Acinetobacter baumannii isolates | Female | Male | ||
| Number | Percentage | Number | Percentage | |||
| Blood | 33 | 30 | 18 | %60 | 12 | %40 |
| Sputum | 45 | 16 | 11 | %68.75 | 5 | %31.25 |
| Urine | 38 | 15 | 6 | %40 | 9 | %60 |
| Trachea | 5 | 1 | 1 | %100 | 0 | 0 |
| Wounds | 19 | 7 | 5 | %71.42 | 2 | %28.57 |
| Pus | 25 | 4 | 3 | %75 | 1 | %25 |
Phenotypic Evaluation of Antibiotic Resistance Pattern
Phenotypic evaluation of the antibiotic resistance patterns among A. baumannii strains indicated that the highest resistance was related to ceftazidime (93.15%) and imipenem (94.52%) and the lowest resistance was related to amikacin (47.94%). The antibiotic resistance pattern of A. baumannii isolate is given in Table 4.
Table 4. Antibiotic susceptibility pattern of A. baumannii isolates
| Antibiotics |
Drug concentration | Number of sensitive strains | Number of intermediate strains | Number of resistant strains |
| Frequency (percentage) | Frequency (percentage) | Frequency (percentage) | ||
| Tobramycin | 10μg | 17(23.28) | 4(5.84) | 52(71.23) |
| Cefepime | 30μg | 10(13.7) | 0(0) | 63(86/30) |
| Imipenem | 10μg | 2(2.73) | 2(2.73) | 69(94/52) |
| Meropenem | 10μg | 6(8.21) | 0(0) | 67(91/78) |
| Amikacin | 30μg | 24(32.88) | 14(19.17) | 35(47/95) |
| Gentamicin | 30μg | 11(15.06) | 3(4.10) | 59(80.82) |
| Ciprofloxacin | 5μg | 8(10.96) | 19(26.02) | 46(63.01) |
| Cefotaxime | 30μg | 5(6.84) | 3(4.10) | 65(89.04) |
| Ceftazidime |
30μg | 3(4/10) | 2(2.73) | 68(93.15) |
Polymerase Chain Reaction Test Results for blaOXA-51 and bap Genes
The results of the polymerase chain reaction showed that the abundance of the blaOXA-51 gene was traced in all 73 (100%) A .baumannii isolates. Also, the frequency of the bap gene was 39 isolates (53.42%). Correlation between the presence of bap genes and the ability to produce biofilm showed that from 73 studied isolates, 8 isolates with multiple drug resistance were able to form strong biofilm. Overall, the PCR results were consistent with the both TM and TCP methods. Four isolates of the blood samples by the PCR method had the bap gene, but they were reported negative in phenotypic biofilm methods. The correlation of them was investigated using SPSS 22 statistical software and t-test, and the ability to form biofilm in resistant isolates was significantly higher than the sensitive isolates (P<0.05).
Assessment of Biofilm Formation Through the Tubular Method
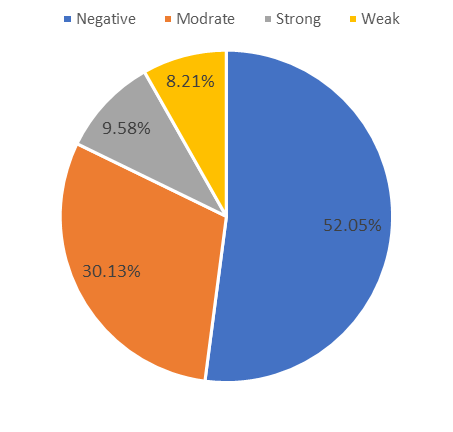
The results of tubular biofilm production depicted that from the 73 studied isolates in this study, 38 isolates (52.05%) were negative, 6 isolates (8.21%) were weak, 22 isolates (30.13%) were moderate, and 7 isolates (9.58%) were classified as strong biofilm producer.
Figure 1. Electrophoresis of PCR products to search for blaOXA-51 gene.
From the left side, the wells include negative control (C-), Ladder, and positive control (C +), (A. baumannii ATCC 19606), respectively. The wells 1 to 6 are blood isolates, 7 is pus isolate, wells 8, 9, 10 are urine isolates, and wells are 11, 12 sputum isolates.
Figure 2. Electrophoresis of PCR products to search for the bap gene. The wells from the right side include the negative control (C-), Ladder and positive control (C +) (A. baumannii ATCC 19606) and the isolates (wells 1 to 5 include blood isolates), respectively.
Evaluation of Biofilm Formation using a Microtiter Plate Test Method
The study results of microtiter plate production biofilm indicated that from 73 A. baumannii isolates, 35 isolates (47.94%) were negative, 5 isolates (6.84%) were weak, 25 isolates (34.24%) were medium and 8 isolates (10.95%) were classified as strong biofilm producer. The results of both tubular and microtiter plate methods are given comparatively in Chart 1 and 2.
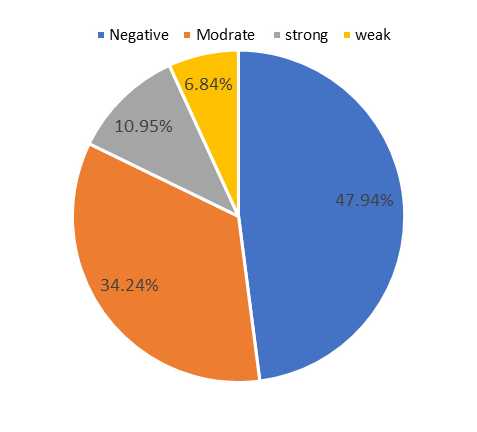
Chart 1. The rate of biofilm formation by a tubular method
Chart 2. Biofilm formation by microtiter plate method
Amount of Biofilm Production in Plate Microtiter Method Based on Drug Resistance
The comparison rate of biofilm formation in the studied strains was done by a quantitative microtiter plate method with the rate of resistance and susceptibility of antibiotic (Figure 3). The results demonstrated that the rate of biofilm formation in antibiotic resistant strains is higher than antibiotic susceptible strains.
Figure 3. Comparison of drug resistance and drug sensitivity with the rate of biofilm production based on the microtiter plate method
| Sample isolation place | Number of biofilm formation (percentage) | Presence of bap gene in isolated strains (percentage) | ||||
| Tubular | Microtitr plate | |||||
| bap | ||||||
| + | - | + | - | |||
| Blood | 11(36.66) | 19(63.66) | 12(40) | 18(60) | 12(%40) | |
| Sputum | 10(62.5) | 6(37.5) | 9(56.25) | 7(43.75) | 10(%62.5) | |
| Urine | 8(53.33) | 7(46.66) | 9(60) | 6(40) | 9(%60) | |
| Trachea | 0(0) | 1(100) | 0(0) | 1(100) | 0(0) | |
| Wounds | 4(57/14) | 3(42.85) | 5(71.42) | 2(28.57) | 5(%71.42) | |
| Pus | 2(50) | 2(50) | 3(75) | 1(25) | 3(%75) | |
| Total | 35(47.94) | 38(52.05) | 38(52.5) | 35(47.94) | 39(%53.42) | |
Discussion
Due to the increasing drug resistance to different antibiotics during recent years, we are witness to the isolates of A. baumannii with multiple drug resistance pattern (MDR) that are reported from around the world (17). Increasing resistance to antimicrobial agents and limited treatment options are serious problems in medical centers, especially in the intensive care unit (ICU), which affects the health and economy of the country (18). The ability to attach and form biofilm on mucus and medical devices is one of the main factors of virulence and drug resistance in Acinetobacter species. Bacterial biofilm causes the bacterium resistant to factors such as the presence of antibiotics, host immune response, phagocytic agents, and environmental stress (11).
The study of the biofilm formation ability and the detection of genes involved in various Acinetobacter isolates can help a better understanding of the process of biofilm formation. Consequently, one of the important purposes of this study was to investigate the rate of biofilm formation in the antibiotic-resistant clinical A. baumannii strains. The present study revealed that most isolates of A. baumannii have a multiple drug resistance pattern. The highest resistance to imipenem (94.52%) and the lowest resistance to amikacin (47.95%) were evaluated.
The presence of A. baumannii isolates with multiple drug resistance patterns has been reported in other studies in Iran. In the study of Hatami et al. in 2018, 84% of A. baumannii isolates represented multiple drug resistance (19). In a study that conducted by Goodarzi and colleagues in 2013, resistance to ceftazidime and meropenem was 99% and to imipenem was 91.5% (20), and in the study of Kooti et al. in 2015, resistance to ceftazidime and meropenem was 99.5% and to imipenem was 98.5% (1). Also, in the study of Maraki et al. conducted in Greece in 2015, the rate of resistance to cefotaxime and gentamicin was reported 96.2% and 91.5%, respectively (21), all of them indicate a very high resistance of this bacterium to a wide range of different antibiotics. In general, the rate of antibiotic resistance is higher in Asian and European countries than in the United States and Latin America, and this rate of resistance in Iran is very alarming (22). Many studies demonstrate the survival power of A. baumannii in harsh environmental conditions and very high resistance to various antibiotics caused by biofilm formation (23). The bap gene is one of the most important generating and amplifying genes of biofilm in A. baumannii. This protein with high molecular weight is located on the outer surface of bacteria and includes a central core consisting of sequential repeats of similar sequences (9). In recent years, many researchers have studied the relationship between A. baumannii antibiotic resistance and involved genes in the biofilm production of this bacterium. Lihao Ki and colleagues conducted a study in 2016 on the association between antibiotic resistance, biofilm formation, and biofilm specific resistance in A. baumannii. In their study, the association between antibiotic resistance, biofilm formation, and biofilm specific resistance in clinical isolates of A.baumannii was assessed (24).
In a study by Ji Yong Song et al. in 2016, from 92 isolates of multidrug-resistant A. baumannii, all strains contained the bap gene (25). Also, in similar studies conducted in Australia and the United States in 2013 and 2015, the abundance of this gene was reported 92% and 84%, respectively (26,27). The frequency of the bap gene in the present study was 53.42%, which despite the mismatch between the prevalence of this gene in the studies outside of Iran with the present study, can be due to the different geographical conditions, number, or variety of samples. In the current study also, all A. baumannii isolates contained the blaOXA-51 gene. In studies conducted by Amani et al. in 2018 and Hedayati et al. in 2019, they also reported a 100% abundance of blaOXA-51 . Due to the existence of this gene in all isolates of A. baumannii, tracking of the gene is the quickest way to detect this bacterium (28, 29).
In order to evaluation of biofilm formation ability, the plate microtiter method was used. Additionally, the microtiter plate method detects the biofilm formation ability of isolates; it can measure the rate of biofilm formation quantitatively. In the current study by this method, 52.6% of the isolates formed biofilm and 47.94% of them were negative in biofilm formation. These results were similar to the findings of El Khair et al. In their study in 2015, 51.3% of the isolates were biofilm positive and 48.7% of the isolates were biofilm negative (30). The second was the tubular method, which used to evaluate the biofilm formation ability of A. baumannii. This method was able to identify biofilm in 47.95% of isolates also, 52.05% of the isolates were negative in biofilm formation, which was slightly different from the obtained results from the plate microtiter test. In this study, all isolates producing strong and moderate biofilm generally were contained bap gene. Considering the low frequency of the ability to form strong biofilm in the studied isolates, it is probable that other reasons such as changes in penicillin-binding proteins, decreased membrane permeability, production of antibiotic hydrolyzing enzymes, or expression of efflux pumps cause this amount of resistance.
Research Limitations: The lack of collaboration of the referral patients in completing the questionnaires and the probability of using antibiotics by the patient were the limitations of this study.
Conclusion
In the present study, the resistance rate of isolates was evaluated by a disk diffusion method and the correlation between resistant isolates with biofilm formation was compared, which indicating the formation of biofilm by most resistant isolates. Also, the frequency of the bap gene had a maximum consistent with the number of isolates forming biofilm. However, the bap gene is not the only gene involved in biofilm formation, but it can be said that it has a significant role in the formation of biofilm by this bacterium.
Acknowledgements
The authors thank all those who helped them writing this article.
Conflicts of Interest
Authors declared no conflict of interests.
Ethical considerations
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Funding and support
This research resulted from an independent research without receiving any financial support.
Received: 2020/04/22 | Accepted: 2020/08/17 | ePublished: 2020/10/27
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |